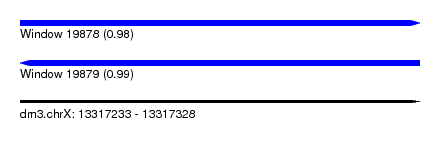
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,317,233 – 13,317,328 |
| Length | 95 |
| Max. P | 0.987531 |

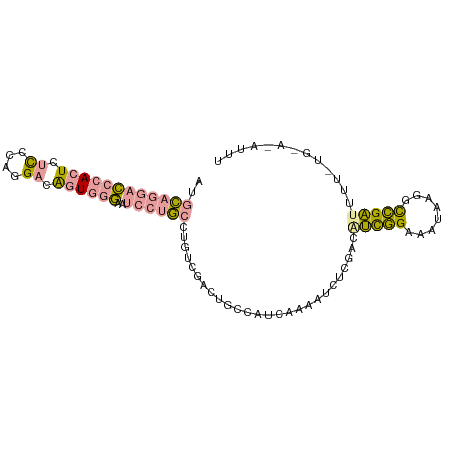
| Location | 13,317,233 – 13,317,328 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 52.28 |
| Shannon entropy | 0.67041 |
| G+C content | 0.43179 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -12.43 |
| Energy contribution | -14.00 |
| Covariance contribution | 1.57 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.981068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13317233 95 + 22422827 AAAUGUACACAAAAACUAAUUUCUUGCAAUCCGCUUAAAAGUGAUAGGACUAGUUAGCAUUGGUAAUUAGCUGCUCAAAACAGUUUUUAUUUUAU ..........(((((((..(((...((....((((....))))...((.(((((((.(...).)))))))))))..)))..)))))))....... ( -12.80, z-score = 0.79, R) >dp4.chr4_group1 4225536 95 + 5278887 AAAUUUUCAAAAAAUCGGCCUCAUUUCCGAUGUCGAGAUUUUGAUGGCAGUCGACAGGCAGGAUCCCCACUGUCCUGGGAGAGUGGGUCCUGCAU ......((((((..(((((.((......)).)))))..)))))).............((((((..((((((.((....)).)))))))))))).. ( -37.00, z-score = -2.91, R) >droPer1.super_5 5774457 85 - 6813705 ----------AAAAUCGGCCUUAUUUCCGAAGUGGAGAUUUUUAUGGCAAUCGACAGGCAGGAUUCCCACUGUCCUGGGAGAGUGGGUCCUGCAC ----------....((((((..((((((.....))))))......)))...)))...((((((..((((((.((....)).)))))))))))).. ( -34.00, z-score = -3.09, R) >consensus AAAU_U_CA_AAAAUCGGCCUUAUUUCCGAAGUCGAGAUUUUGAUGGCAAUCGACAGGCAGGAUACCCACUGUCCUGGGAGAGUGGGUCCUGCAU ..............((((((..(((((.......)))))......)))...)))...((((((..((((((.((....)).)))))))))))).. (-12.43 = -14.00 + 1.57)

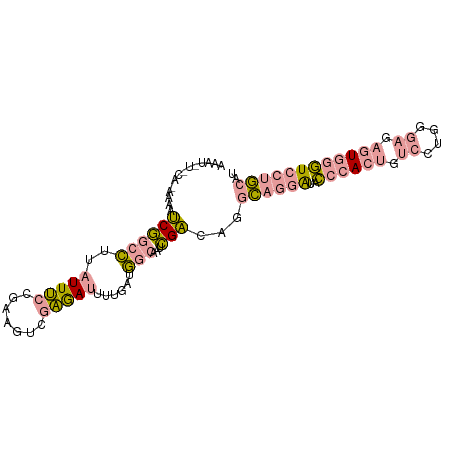
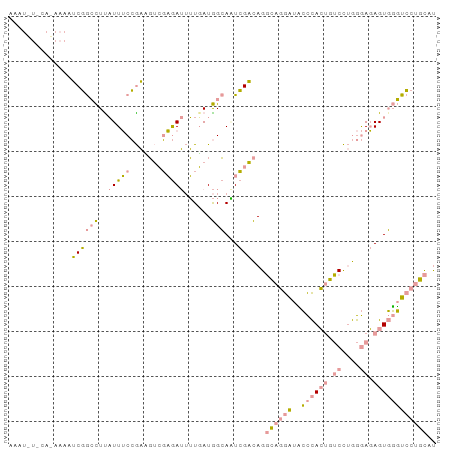
| Location | 13,317,233 – 13,317,328 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 52.28 |
| Shannon entropy | 0.67041 |
| G+C content | 0.43179 |
| Mean single sequence MFE | -25.11 |
| Consensus MFE | -12.63 |
| Energy contribution | -13.20 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.987531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13317233 95 - 22422827 AUAAAAUAAAAACUGUUUUGAGCAGCUAAUUACCAAUGCUAACUAGUCCUAUCACUUUUAAGCGGAUUGCAAGAAAUUAGUUUUUGUGUACAUUU .....((((((((((((((..(((((...........)))....(((((..............)))))))..)))).))))))))))........ ( -16.04, z-score = -0.38, R) >dp4.chr4_group1 4225536 95 - 5278887 AUGCAGGACCCACUCUCCCAGGACAGUGGGGAUCCUGCCUGUCGACUGCCAUCAAAAUCUCGACAUCGGAAAUGAGGCCGAUUUUUUGAAAAUUU ..((((((((((((.((....)).))))))..))))))..((((...(((.(((...((.((....))))..))))))))))............. ( -31.20, z-score = -1.96, R) >droPer1.super_5 5774457 85 + 6813705 GUGCAGGACCCACUCUCCCAGGACAGUGGGAAUCCUGCCUGUCGAUUGCCAUAAAAAUCUCCACUUCGGAAAUAAGGCCGAUUUU---------- ..((((((((((((.((....)).))))))..)))))).((..((((........))))..))..((((........))))....---------- ( -28.10, z-score = -2.51, R) >consensus AUGCAGGACCCACUCUCCCAGGACAGUGGGAAUCCUGCCUGUCGACUGCCAUCAAAAUCUCGACAUCGGAAAUAAGGCCGAUUUU_UG_A_AUUU ..((((((((((((.((....)).))))))..))))))..........................(((((........)))))............. (-12.63 = -13.20 + 0.57)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:40:10 2011