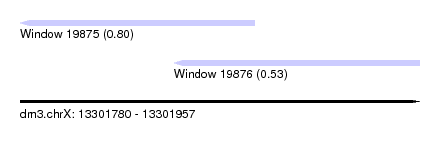
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,301,780 – 13,301,957 |
| Length | 177 |
| Max. P | 0.800529 |

| Location | 13,301,780 – 13,301,884 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 88.46 |
| Shannon entropy | 0.16293 |
| G+C content | 0.46056 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -26.01 |
| Energy contribution | -27.23 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.800529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13301780 104 - 22422827 AAUGUGGAACUUAAUGCCCUCAAAAGAGGGGCGUGUCUGGCCGGCGAAAGUCAAAGUCAUUGUCAAUGUCGCCUGUUUGGAGCCAUUUAGUUUCUGUUUAGGUU .....((((((.(((((((((....)))))((...((..((.(((((..(.(((.....))).)....))))).))..)).)))))).)))))).......... ( -29.80, z-score = -1.18, R) >droSec1.super_21 1054188 98 - 1102487 AAUGUAGGAAUUAAUGCCCUCAAAAGGGGGGCUUGUCUGGCCGGCGAAAGGCAAAGUCAUUGUCAAUGUCUCCUGUUUGGAGUGUUUUAGUUUCUGUU------ ....((((((((((.(((..((((.((((((((((.(((((.(.(....).)...))))..).))).))))))).))))..).)).))))))))))..------ ( -32.10, z-score = -2.64, R) >droSim1.chrX 10194062 104 - 17042790 AAUGUAGGAAUUAAUGCCCUCAAAAGGGGGGCGUGUCUGGCCGGCGAAAGGCAAAGUCAUUGUCAAUGUCGCCUGUUUGGAGUGUUUUAGUCUCUGUUUCUGUU ....(((..((((((((((((.....)))))))).((..((.(((((..(((((.....)))))....))))).).)..))......))))..)))........ ( -31.70, z-score = -1.86, R) >consensus AAUGUAGGAAUUAAUGCCCUCAAAAGGGGGGCGUGUCUGGCCGGCGAAAGGCAAAGUCAUUGUCAAUGUCGCCUGUUUGGAGUGUUUUAGUUUCUGUUU__GUU ....((((((((((.((((((.....))))))...((..((.(((((..(((((.....)))))....))))).).)..)).....))))))))))........ (-26.01 = -27.23 + 1.23)

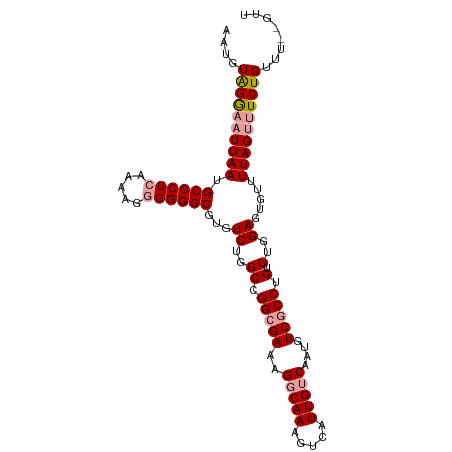
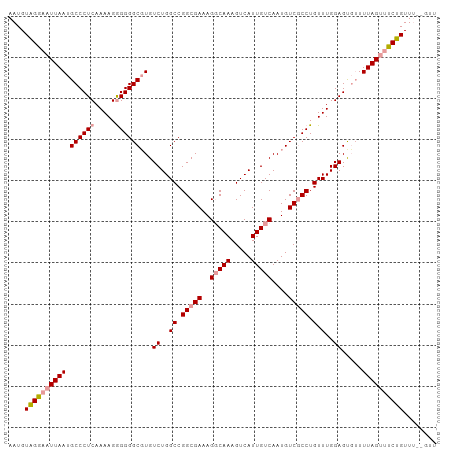
| Location | 13,301,848 – 13,301,957 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 67.92 |
| Shannon entropy | 0.60401 |
| G+C content | 0.45494 |
| Mean single sequence MFE | -25.85 |
| Consensus MFE | -9.91 |
| Energy contribution | -10.47 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.529014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
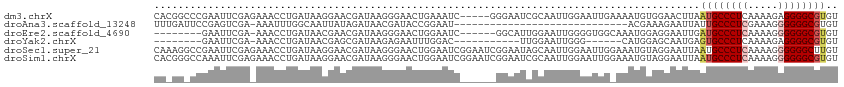
>dm3.chrX 13301848 109 - 22422827 CACGGCCCGAAUUCGAGAAACCUGAUAAGGAACGAUAAGGGAACUGAAAUC-----GGGAAUCGCAAUUGGAAUUGAAAAUGUGGAACUUAAUGCCCUCAAAAGAGGGGCGUGU ((((.(((((.(((......(((....))).......((....))))).))-----)))..(((((.((........)).))))).........(((((....))))).)))). ( -30.50, z-score = -2.07, R) >droAna3.scaffold_13248 1269678 84 - 4840945 UUUGAUUCCGAGUCGA-AAAUUUGGCAAUUAUAGAUAACGAUACCGGAAU-----------------------------ACGAAAGAAUUAUUGCCCUCGAAAGGGGGGCGUGU ((((((((((.((((.-..(((((.......)))))..))))..))))))-----------------------------.))))..........(((((....)))))...... ( -23.10, z-score = -1.89, R) >droEre2.scaffold_4690 5180162 99 + 18748788 --------GAAUUCGA-AAACCUGAUAACGAACGAUAAGGGAACUGGAAUC------GGCAUUGGAAUUGGGGUGGCAAAUGGAGGAAUUGAUGCCCUCAAAAGGGGGGCGUGU --------.(((((..-...((..((..(((.((((.((....))...)))------)...)))..))..))((.....))....))))).((((((((.....)))))))).. ( -26.90, z-score = -1.88, R) >droYak2.chrX 7580980 88 - 21770863 --------GAAUUCGA-AAACCUGAUAACGAGCGAUAAGAGAAUUUGGAC-----------UUGGAAUUGGG------CAUGGAGCAAUGAGUGCCCUCAAAAGAGGGGCGUGU --------........-...((..((..(((((.(..........).).)-----------)))..))..))------((((..((.....)).(((((....))))).)))). ( -18.70, z-score = -0.26, R) >droSec1.super_21 1054250 114 - 1102487 CAAAGGCCGAAUUCGAGAAACCUGAUAAGGAACGAUAAGGGAACUGGAAUCGGAAUCGGAAUAGCAAUUGGAAUUGGAAAUGUAGGAAUUAAUGCCCUCAAAAGGGGGGCUUGU ......((((.(((......(((....)))..((((.((....))...))))))))))).....((((....)))).................((((((.....)))))).... ( -25.90, z-score = -0.87, R) >droSim1.chrX 10194130 114 - 17042790 CACGGGCCAAAUUCGAGAAACCUGAUAAGGAACGAUAAGGGAACUGGAAUCGGAAUCGGAAUCGCAAUUGGAAUUGGAAAUGUAGGAAUUAAUGCCCUCAAAAGGGGGGCGUGU .(((..((((.(((((....(((....)))..((((.((....))...))))...............))))).))))...)))........((((((((.....)))))))).. ( -30.00, z-score = -1.44, R) >consensus CA_GG_CCGAAUUCGA_AAACCUGAUAAGGAACGAUAAGGGAACUGGAAUC______GGAAUCGCAAUUGGAAUUG_AAAUGUAGGAAUUAAUGCCCUCAAAAGGGGGGCGUGU ...........................................................................................((((((((.....)))))))).. ( -9.91 = -10.47 + 0.56)
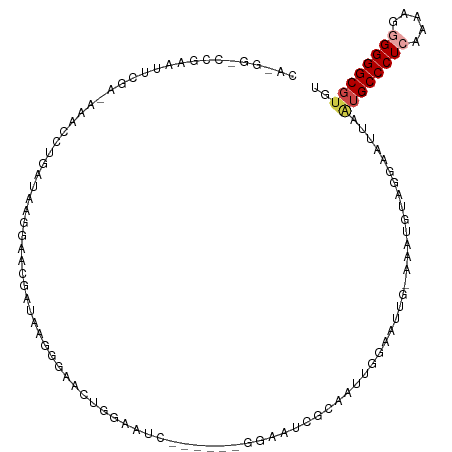
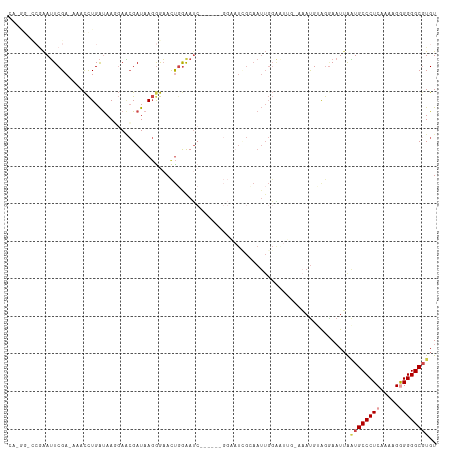
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:40:08 2011