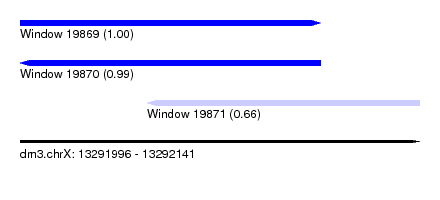
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,291,996 – 13,292,141 |
| Length | 145 |
| Max. P | 0.996155 |

| Location | 13,291,996 – 13,292,105 |
|---|---|
| Length | 109 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 70.24 |
| Shannon entropy | 0.54834 |
| G+C content | 0.44254 |
| Mean single sequence MFE | -35.24 |
| Consensus MFE | -16.87 |
| Energy contribution | -16.19 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.996155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
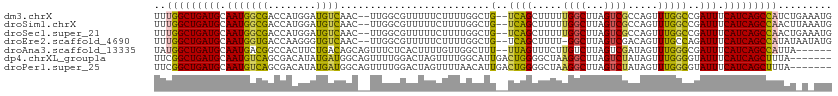
>dm3.chrX 13291996 109 + 22422827 UUUGGCUGAUGCAAUGGCGACCAUGGAUGUCAAC--UUGGCGUUUUUCUUUUGGCUG--UCAGCUUUUUGGCUUAGUCGCCAGUUUGGCCGAUUUCAUCAGCCAUCUGAAAUG ..(((((((((.(((((((.(((.(((((((...--..)))))))......))).))--)).(((..(((((......)))))...)))..))).)))))))))......... ( -36.90, z-score = -2.24, R) >droSim1.chrX 10185357 109 + 17042790 UUUGGCUGAUGCAAUGGCGACCAUGGAUGUCAAC--UUGGCGUUUUUCUUUUGGCUG--UCAGCUUUUUGGCUUAGUCGCCAGUUUGGCCGAUUUCAUCAGCCAACUUAAAUG .((((((((((.(((((((.(((.(((((((...--..)))))))......))).))--)).(((..(((((......)))))...)))..))).))))))))))........ ( -37.70, z-score = -2.85, R) >droSec1.super_21 1045277 109 + 1102487 UUUGGCUGAUGCAAUGGCGACCAUGGAUGUCAAC--UUGGCGUUUUUCUUUUGGCUG--UCAGCUUUUUGGCUUAGUCGCCAGUUUGGCCGAUUUCAUCAGCCAACUGAAAUG .((((((((((.(((((((.(((.(((((((...--..)))))))......))).))--)).(((..(((((......)))))...)))..))).))))))))))........ ( -37.70, z-score = -2.50, R) >droEre2.scaffold_4690 5172155 108 - 18748788 UUUGGCUGAUGCAAUGGUGACCAAGGGUGUCAAC--UUGGCGUUUUUCUUUUGGCUG--UCAGCUUUU-GGCUUAGUCGACAGUUUGCCAGAUUUCAUCAGCCAUAUAAUAUG ..(((((((((...(((..((((((........)--)))).............((((--((.(((...-.....))).)))))))..))).....)))))))))......... ( -37.00, z-score = -3.04, R) >droAna3.scaffold_13335 2492607 105 - 3335858 UAUGGCUGAUGCAAUGACGGCCACUUCUGACAGCAGUUUCUCACUUUUGUUGGCUUU--UUAGUUUCUUGUCUUAGUCGAUAGUUUGGGCGAUUUCAUCAGCCAUUA------ .((((((((((.(((..((.(((...(..((((.(((.....))).))))..)....--........(((((......)))))..))).))))).))))))))))..------ ( -30.20, z-score = -2.27, R) >dp4.chrXL_group1a 3191090 106 - 9151740 UUCGGCUGAUGCAAUGUCAGCGACAUAUGAUGGCAGUUUUGGACUAGUUUUGGCAUUGACUGGGGCUAAGGCUUAGUCUAUAGUUUGGGGUAUUUCAUCAGCUUUA------- ...((((((((.(((..(...((((((.(((...((((((((.((((((........))))))..))))))))..)))))).)))...)..))).))))))))...------- ( -33.60, z-score = -2.54, R) >droPer1.super_25 1413497 106 + 1448063 UUCGGCUGAUGCAAUGUCAGCGACAUAUGAUGGCAGUUUUGGACUAGUUUUAACAUUGACUGGGGCUAAGGCUUAGUCUAUAGUUUGGGGUAUUUCAUCAGCUUUA------- ...((((((((.(((..(...((((((.(((...((((((((.((((((........))))))..))))))))..)))))).)))...)..))).))))))))...------- ( -33.60, z-score = -3.10, R) >consensus UUUGGCUGAUGCAAUGGCGACCACGGAUGUCAAC__UUGGCGUUUUUCUUUUGGCUG__UCAGCUUUUUGGCUUAGUCGACAGUUUGGCCGAUUUCAUCAGCCAUAU_A_AUG ..(((((((((((((((((((..(((...))).....................(((.....)))...........)))))))..)))........)))))))))......... (-16.87 = -16.19 + -0.69)
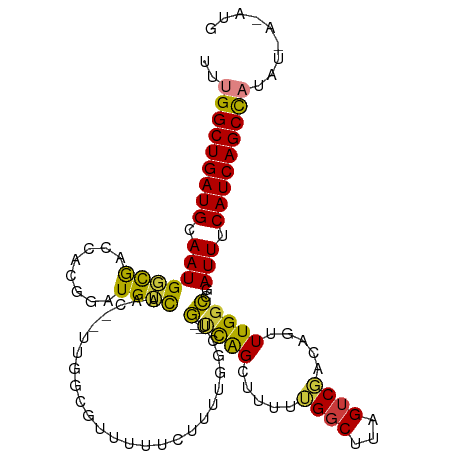
| Location | 13,291,996 – 13,292,105 |
|---|---|
| Length | 109 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 70.24 |
| Shannon entropy | 0.54834 |
| G+C content | 0.44254 |
| Mean single sequence MFE | -27.86 |
| Consensus MFE | -11.11 |
| Energy contribution | -11.93 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.987537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13291996 109 - 22422827 CAUUUCAGAUGGCUGAUGAAAUCGGCCAAACUGGCGACUAAGCCAAAAAGCUGA--CAGCCAAAAGAAAAACGCCAA--GUUGACAUCCAUGGUCGCCAUUGCAUCAGCCAAA .........(((((((((.(((((((((...((((.....(((......)))..--..)))).......(((.....--)))........))))))..))).))))))))).. ( -33.70, z-score = -2.98, R) >droSim1.chrX 10185357 109 - 17042790 CAUUUAAGUUGGCUGAUGAAAUCGGCCAAACUGGCGACUAAGCCAAAAAGCUGA--CAGCCAAAAGAAAAACGCCAA--GUUGACAUCCAUGGUCGCCAUUGCAUCAGCCAAA ........((((((((((.(((((((((...((((.....(((......)))..--..)))).......(((.....--)))........))))))..))).)))))))))). ( -34.50, z-score = -3.29, R) >droSec1.super_21 1045277 109 - 1102487 CAUUUCAGUUGGCUGAUGAAAUCGGCCAAACUGGCGACUAAGCCAAAAAGCUGA--CAGCCAAAAGAAAAACGCCAA--GUUGACAUCCAUGGUCGCCAUUGCAUCAGCCAAA ........((((((((((.(((((((((...((((.....(((......)))..--..)))).......(((.....--)))........))))))..))).)))))))))). ( -34.50, z-score = -3.03, R) >droEre2.scaffold_4690 5172155 108 + 18748788 CAUAUUAUAUGGCUGAUGAAAUCUGGCAAACUGUCGACUAAGCC-AAAAGCUGA--CAGCCAAAAGAAAAACGCCAA--GUUGACACCCUUGGUCACCAUUGCAUCAGCCAAA .........(((((((((.....(((....((((((.((.....-...)).)))--))).............(((((--(........))))))..)))...))))))))).. ( -31.20, z-score = -3.47, R) >droAna3.scaffold_13335 2492607 105 + 3335858 ------UAAUGGCUGAUGAAAUCGCCCAAACUAUCGACUAAGACAAGAAACUAA--AAAGCCAACAAAAGUGAGAAACUGCUGUCAGAAGUGGCCGUCAUUGCAUCAGCCAUA ------..((((((((((.(((.((........((......))...........--...(((((((..(((.....)))..)))......)))).)).))).)))))))))). ( -25.10, z-score = -2.42, R) >dp4.chrXL_group1a 3191090 106 + 9151740 -------UAAAGCUGAUGAAAUACCCCAAACUAUAGACUAAGCCUUAGCCCCAGUCAAUGCCAAAACUAGUCCAAAACUGCCAUCAUAUGUCGCUGACAUUGCAUCAGCCGAA -------....(((((((.................((((..((....))...))))...............................(((((...)))))..))))))).... ( -17.20, z-score = -1.53, R) >droPer1.super_25 1413497 106 - 1448063 -------UAAAGCUGAUGAAAUACCCCAAACUAUAGACUAAGCCUUAGCCCCAGUCAAUGUUAAAACUAGUCCAAAACUGCCAUCAUAUGUCGCUGACAUUGCAUCAGCCGAA -------....(((((((.................((((..((....))...))))(((((((.....(((.....)))((.((.....)).))))))))).))))))).... ( -18.80, z-score = -2.10, R) >consensus CAU_U_AGAUGGCUGAUGAAAUCGGCCAAACUGUCGACUAAGCCAAAAAGCUGA__CAGCCAAAAGAAAAACGCCAA__GUUGACAUCCAUGGUCGCCAUUGCAUCAGCCAAA .........(((((((((..............(((.....((........))......)))............................(((.....)))..))))))))).. (-11.11 = -11.93 + 0.82)

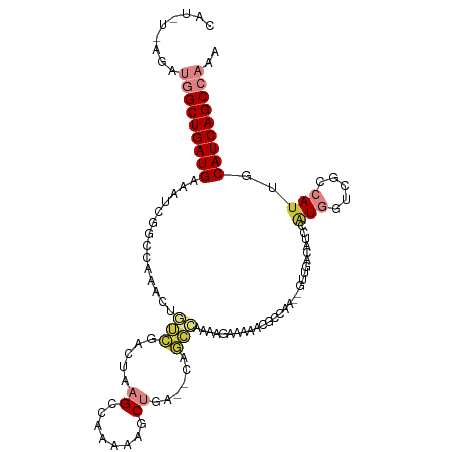
| Location | 13,292,042 – 13,292,141 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 72.87 |
| Shannon entropy | 0.49845 |
| G+C content | 0.38962 |
| Mean single sequence MFE | -22.48 |
| Consensus MFE | -10.09 |
| Energy contribution | -10.87 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.659102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
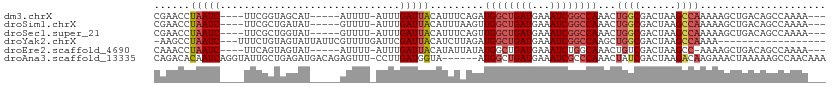
>dm3.chrX 13292042 99 - 22422827 CGAACCUAAUC----UUCGGUAGCAU-----AUUUU-AUUUGAUUACAUUUCAGAUGGCUGAUGAAAUCGGCCAAACUGGCGACUAAGCCAAAAAGCUGACAGCCAAAA--- ...(((.....----...))).....-----.....-((((((.......))))))(((((......(((((.....((((......))))....))))))))))....--- ( -23.90, z-score = -1.93, R) >droSim1.chrX 10185403 99 - 17042790 CGAACCUAAUC----UUCGCUGAUAU-----GUUUU-AUUUGAUUACAUUUAAGUUGGCUGAUGAAAUCGGCCAAACUGGCGACUAAGCCAAAAAGCUGACAGCCAAAA--- ...........----...((((.((.-----(((((-.................(((((((((...)))))))))..((((......)))).))))))).)))).....--- ( -24.70, z-score = -2.51, R) >droSec1.super_21 1045323 99 - 1102487 CGAACCUAAUC----UUCGCUGGUAU-----GUUUU-AUUUGAUUACAUUUCAGUUGGCUGAUGAAAUCGGCCAAACUGGCGACUAAGCCAAAAAGCUGACAGCCAAAA--- ...........----...(((((((.-----(((..-....))))))...(((((((((((((...)))))))....((((......))))...)))))))))).....--- ( -27.00, z-score = -2.40, R) >droYak2.chrX 7572931 90 - 21770863 -AAGCCUAAUC---UUUCUGUAGUAUUAUUCGUUUUGAUUCGAUUACAUCUUAGAUGGCUGAUGAAAUCGGCCAAGCUGGCGACUAAGCCAAAA------------------ -.(((...(((---(...((((((...(((......)))...))))))....))))(((((((...)))))))..)))(((......)))....------------------ ( -22.50, z-score = -1.66, R) >droEre2.scaffold_4690 5172201 98 + 18748788 CAAACCUAAUC----UUCAGUAGUAU-----AUUUU-AUUUGAUUACAUAUUAUAUGGCUGAUGAAAUCUGGCAAACUGUCGACUAAGCC-AAAAGCUGACAGCCAAAA--- .........((----.(((((.((((-----(...(-((........))).))))).))))).))....((((....(((((.((.....-...)).)))))))))...--- ( -17.70, z-score = -0.95, R) >droAna3.scaffold_13335 2492652 105 + 3335858 CAGACACAAUCAGGUAUUGCUGAGAUGACAGAGUUU-CCUUGAUGGUA------AUGGCUGAUGAAAUCGCCCAAACUAUCGACUAAGACAAGAAACUAAAAAGCCAACAAA ............(((....(((......)))(((((-(.((((((((.------..(((.(((...))))))...)))))))).........)))))).....)))...... ( -19.10, z-score = -0.57, R) >consensus CAAACCUAAUC____UUCGCUAGUAU_____AUUUU_AUUUGAUUACAUUUCAGAUGGCUGAUGAAAUCGGCCAAACUGGCGACUAAGCCAAAAAGCUGACAGCCAAAA___ .......................................................((((((((...))))))))...((((......))))..................... (-10.09 = -10.87 + 0.78)
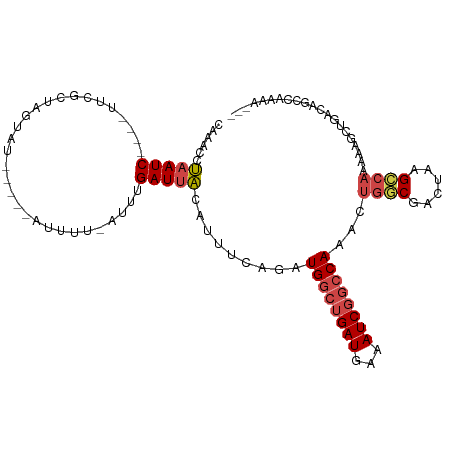
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:40:04 2011