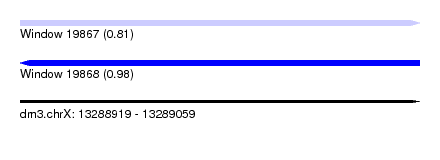
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,288,919 – 13,289,059 |
| Length | 140 |
| Max. P | 0.984883 |

| Location | 13,288,919 – 13,289,059 |
|---|---|
| Length | 140 |
| Sequences | 3 |
| Columns | 143 |
| Reading direction | forward |
| Mean pairwise identity | 84.83 |
| Shannon entropy | 0.20840 |
| G+C content | 0.54321 |
| Mean single sequence MFE | -44.93 |
| Consensus MFE | -37.92 |
| Energy contribution | -37.73 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
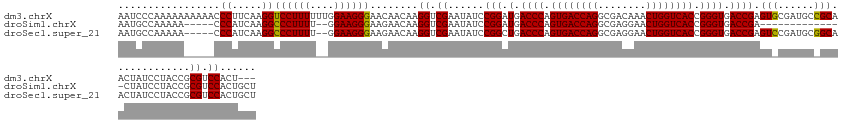
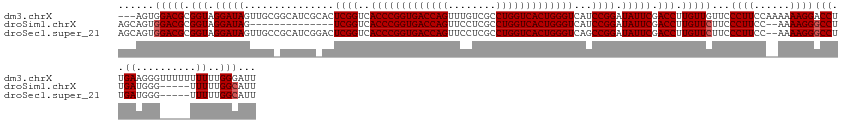
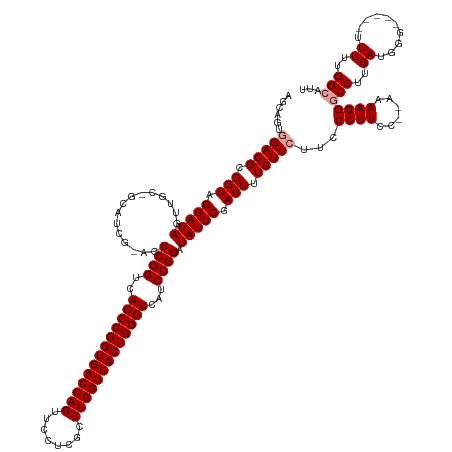
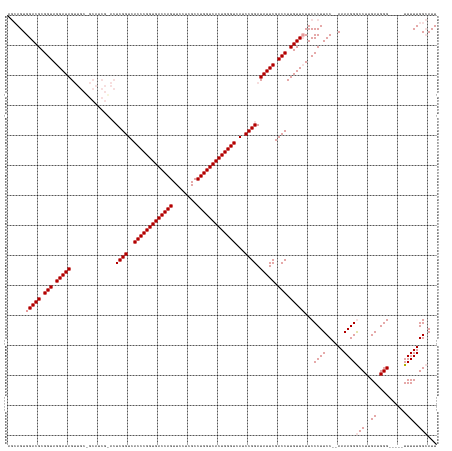
>dm3.chrX 13288919 140 + 22422827 AAUCCCAAAAAAAAAACCCUUCAAGGUCCUUUUUUGGAAGGGAACAACAAGGUCGAAUAUCCGGAUGACCCAGUGACCAGGCGACAAACUGGUCACCGGGUGACCGAGUGCGAUGCCGCAACUAUCCUACCGCGUCCACU--- ..((((.((((((..(((......)))..))))))....)))).......((.((......(((.(.((((.((((((((........)))))))).)))).))))((((((....))).))).........)).))...--- ( -47.50, z-score = -2.80, R) >droSim1.chrX 10182388 122 + 17042790 AAUGCCAAAAA-----CCCAUCAAGGCCCUUUU--GGAAGGGAAGAACAAGGUCGAAUAUCCGGAUGACCCAGUGACCAGGCGAGGAACUGGUCACCGGGUGACCGA--------------CUAUCCUACCGCGUCCACUGCU ....((((((.-----.((.....))...))))--)).(((((.(.....(((.(.(((..(((.(.((((.((((((((........)))))))).)))).)))).--------------.))).).))).).))).))... ( -40.80, z-score = -1.34, R) >droSec1.super_21 1042301 136 + 1102487 AAUGCCAAAAA-----CCCAUCAAGGCCCUUUU--GGAAGGGAAGAACAAGGUCGAAUAUCCGGCUGACCCAGUGACCAGGCGAGGAACUGGUCACCGGGUGACCGAGUCCGAUGCGGCAACUAUCCUACCGCGUCCACUGCU ...(((.....-----.((.....))(((((..--..)))))........)))........(((.(.((((.((((((((........)))))))).)))).))))(((..(((((((...........))))))).)))... ( -46.50, z-score = -0.97, R) >consensus AAUGCCAAAAA_____CCCAUCAAGGCCCUUUU__GGAAGGGAAGAACAAGGUCGAAUAUCCGGAUGACCCAGUGACCAGGCGAGGAACUGGUCACCGGGUGACCGAGU_CGAUGC_GCAACUAUCCUACCGCGUCCACUGCU .................((.....))((((((....))))))........((.((......(((.(.((((.((((((((........)))))))).)))).)))).(((......))).............)).))...... (-37.92 = -37.73 + -0.19)
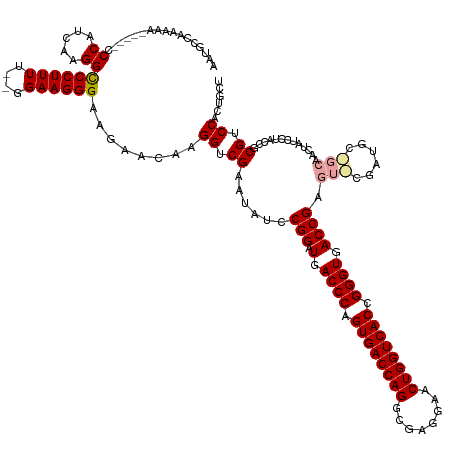
| Location | 13,288,919 – 13,289,059 |
|---|---|
| Length | 140 |
| Sequences | 3 |
| Columns | 143 |
| Reading direction | reverse |
| Mean pairwise identity | 84.83 |
| Shannon entropy | 0.20840 |
| G+C content | 0.54321 |
| Mean single sequence MFE | -55.67 |
| Consensus MFE | -41.72 |
| Energy contribution | -42.39 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.984883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13288919 140 - 22422827 ---AGUGGACGCGGUAGGAUAGUUGCGGCAUCGCACUCGGUCACCCGGUGACCAGUUUGUCGCCUGGUCACUGGGUCAUCCGGAUAUUCGACCUUGUUGUUCCCUUCCAAAAAAGGACCUUGAAGGGUUUUUUUUUUGGGAUU ---....((((.(((.(((((..((((....)))).((((..(((((((((((((........)))))))))))))...)))).))))).))).))))..((((....((((((((((((....)))))))))))).)))).. ( -58.60, z-score = -3.66, R) >droSim1.chrX 10182388 122 - 17042790 AGCAGUGGACGCGGUAGGAUAG--------------UCGGUCACCCGGUGACCAGUUCCUCGCCUGGUCACUGGGUCAUCCGGAUAUUCGACCUUGUUCUUCCCUUCC--AAAAGGGCCUUGAUGGG-----UUUUUGGCAUU .((...(((((.(((.(((((.--------------.(((..(((((((((((((........)))))))))))))...)))..))))).))).))))).........--..((((((((....)))-----))))).))... ( -52.20, z-score = -3.05, R) >droSec1.super_21 1042301 136 - 1102487 AGCAGUGGACGCGGUAGGAUAGUUGCCGCAUCGGACUCGGUCACCCGGUGACCAGUUCCUCGCCUGGUCACUGGGUCAGCCGGAUAUUCGACCUUGUUCUUCCCUUCC--AAAAGGGCCUUGAUGGG-----UUUUUGGCAUU .((..((((.(((((((.....))))))).(((((.(((((.(((((((((((((........)))))))))))))..)))))...)))))..............)))--).((((((((....)))-----))))).))... ( -56.20, z-score = -2.19, R) >consensus AGCAGUGGACGCGGUAGGAUAGUUGC_GCAUCG_ACUCGGUCACCCGGUGACCAGUUCCUCGCCUGGUCACUGGGUCAUCCGGAUAUUCGACCUUGUUCUUCCCUUCC__AAAAGGGCCUUGAUGGG_____UUUUUGGCAUU ......(((((.(((.(((((...............((((..(((((((((((((........)))))))))))))...)))).))))).))).)))))..(((((......)))))((.....))................. (-41.72 = -42.39 + 0.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:40:02 2011