
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,308,154 – 11,308,256 |
| Length | 102 |
| Max. P | 0.871020 |

| Location | 11,308,154 – 11,308,256 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.78 |
| Shannon entropy | 0.34342 |
| G+C content | 0.44516 |
| Mean single sequence MFE | -22.64 |
| Consensus MFE | -11.06 |
| Energy contribution | -11.22 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11308154 102 - 23011544 -----GACAUUCCGACUGACAACAACUCAUAAA-UUUUGCCCACAAAUCGCGC--GGCGAGCG-CACAAAAUCUGUCACCUAUCAAAAAUGCCCUAAAAGGGCAUAAGUCU-------- -----........((((((((............-.((((....))))..((((--.....)))-)........))))...........((((((.....)))))).)))).-------- ( -25.60, z-score = -3.37, R) >droSim1.chr2L 11111892 102 - 22036055 GACAUU-----CCGGCUGACAACAACUCAUAAA-UUUUGCCCACAAAUCGCGC--GGCGAGCG-CACAAAAUCUGUCACCUAUCAAAAAUGCCCUAAAAGGGCAUAAGUCU-------- (((...-----..((.(((((............-.((((....))))..((((--.....)))-)........)))))))........((((((.....))))))..))).-------- ( -28.20, z-score = -3.53, R) >droSec1.super_3 6705172 102 - 7220098 GACAUU-----CCGGCUGACAACAACUCAUAAA-UUUUGCCCACAAAUCGCGC--GGCGAGCG-CACAAAAUCUGUCACCUAUCAAAAAUGCCCUAAAAGGGCAUAAGUUU-------- (((...-----..((.(((((............-.((((....))))..((((--.....)))-)........)))))))........((((((.....))))))..))).-------- ( -25.50, z-score = -2.42, R) >droYak2.chr2L 7705092 103 - 22324452 GACAUUC----CCGGCUGACAACAACUCAUAAA-UUUUGCCCACAAAUCGCGC--CGCGAGCG-CACAAAAUCUGUCACCUAUCAAAAAUGCCCUAAAAGGGCAUAAGUCU-------- (((....----..((.(((((............-.((((....))))..((((--.....)))-)........)))))))........((((((.....))))))..))).-------- ( -27.00, z-score = -3.78, R) >droEre2.scaffold_4929 12506342 107 + 26641161 GACAUUCCAUUCCGGCUGACAACAACUCAUAAA-UUUUGCCCACAAAUCGCGC--CGCGAGCG-CACAAAAUCUGUCACCUAUCAAAAAUGCCCUAAAAGGGCAUAAGUCU-------- (((..........((.(((((............-.((((....))))..((((--.....)))-)........)))))))........((((((.....))))))..))).-------- ( -27.00, z-score = -3.64, R) >droAna3.scaffold_12916 12028605 101 + 16180835 GACAUU-----CUGGCUGACAACAACUCAUAAA-UUUUG-CCACAAAUCGCGC--GGCGAGCG-CACAAAAUCUGUCACCUAUCAAAAAUGCCCUACAAGGGCAUAACUCU-------- ((((..-----.((((.................-....)-)))......((((--.....)))-)........))))...........((((((.....))))))......-------- ( -25.20, z-score = -3.16, R) >dp4.chr4_group3 6744800 97 - 11692001 ----------GACGACUGACAACAACUCAUAAA-UUUUGCCCACAAAUCGCGC--GGCGAGCG-CACAAAAUCUGUCACCUAUCAAAAAUGCCCUAAAAGGGCAUAAGUCU-------- ----------...((((((((............-.((((....))))..((((--.....)))-)........))))...........((((((.....)))))).)))).-------- ( -25.60, z-score = -3.56, R) >droPer1.super_1 8193934 97 - 10282868 ----------GACGACUGACAACAACUCAUAAA-UUUUGCCCACAAAUCGCGC--GGCGAGCG-CACAAAAUCUGUCACCUAUCAAAAAUGCCCUAAAAGGGCAUAAGUCU-------- ----------...((((((((............-.((((....))))..((((--.....)))-)........))))...........((((((.....)))))).)))).-------- ( -25.60, z-score = -3.56, R) >droWil1.scaffold_181038 219677 89 + 637489 --------------------GACAACUCAUAAAUUUUUGCCCACAAAUCGCGC--CGAUACUGCCACAAAAUCUGUCACCUAUCAAAAUUGCCCUAAAAGGGCAUAACUUC-------- --------------------((((...........((((....))))..(((.--......))).........))))............(((((.....))))).......-------- ( -12.60, z-score = -1.47, R) >droVir3.scaffold_12963 17997213 89 + 20206255 --------------------GACGACUCAUAAA-UUUUGUCCACAAAUCGCCG---GCGAGUG-CACAAAAUCUGUCACCUAUCAAAAAUACCCUAAAAGGGCAUAAGUCUGCA----- --------------------((((((......(-((((((.(((...(((...---.))))))-.)))))))..)))..............(((.....))).....)))....----- ( -18.20, z-score = -1.84, R) >droMoj3.scaffold_6500 4942401 94 + 32352404 --------------------GACAACUCAUAAA-UUUUGUCCACAAAUCGCCG---GCGAGUG-CACAAAAUCUGUCACCUAUCAAAAAUACCCUAAAAGGGCAUAAAGUUCUGAAGCA --------------------((((........(-((((((.(((...(((...---.))))))-.))))))).))))..............(((.....)))................. ( -14.40, z-score = -0.40, R) >droGri2.scaffold_15126 7113178 92 + 8399593 --------------------GACAACUCAUAAA-UUGUGUCCACAAAUCGGCGAGUGCGAGUG-CACAAAAUCUGUCACCUAUCAAAAAUACCCUAAAAGGGCAUAAGUCGGCC----- --------------------((((.(.......-..)))))......(((((..((((..(((-.(((.....)))))).............((.....))))))..)))))..----- ( -16.80, z-score = -0.31, R) >consensus ___________CCGGCUGACAACAACUCAUAAA_UUUUGCCCACAAAUCGCGC__GGCGAGCG_CACAAAAUCUGUCACCUAUCAAAAAUGCCCUAAAAGGGCAUAAGUCU________ .....................................(((((.....((((.....)))).....(((.....))).......................)))))............... (-11.06 = -11.22 + 0.17)

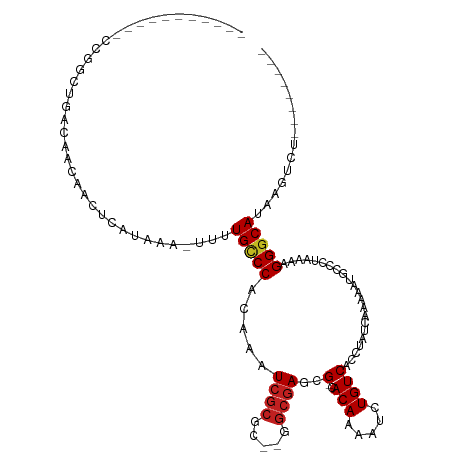

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:03 2011