| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,230,356 – 13,230,447 |
| Length | 91 |
| Max. P | 0.864879 |

| Location | 13,230,356 – 13,230,447 |
|---|---|
| Length | 91 |
| Sequences | 14 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 70.43 |
| Shannon entropy | 0.70089 |
| G+C content | 0.45757 |
| Mean single sequence MFE | -20.76 |
| Consensus MFE | -12.97 |
| Energy contribution | -13.65 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.24 |
| Mean z-score | -0.36 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.864879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13230356 91 - 22422827 -CCUCUUGACUAAUUGUGUUCC---CGAACAUU--UUGCAGCCAUGGUGCGUAUGAACGUAUUGGCCGAUGCCUUGAAGUGCAUAAACAACGCCGAG -...((((.......(((((..---..))))).--..((.((((..((((((....))))))))))..(((((.....).)))).......)))))) ( -18.50, z-score = 0.36, R) >droSim1.chrX 10121682 91 - 17042790 -GCUCUUGACUAAUUGUGUUCC---CGAACAUU--UUGCAGCCAUGGUGCGUAUGAACGUAUUGGCCGAUGCCCUCAAGUGCAUAAACAACGCCGAG -((.(((((......(((((..---..))))).--..(((((((..((((((....))))))))))...)))..))))).))............... ( -22.40, z-score = -0.61, R) >droSec1.super_21 985588 91 - 1102487 -GCUCUUGACUAAUUGUGUUCC---CGAACAUU--UUGCAGCCAUGGUGCGUAUGAACGUAUUGGCCGAUGCCCUCAAGUGCAUAAACAACGCCGAG -((.(((((......(((((..---..))))).--..(((((((..((((((....))))))))))...)))..))))).))............... ( -22.40, z-score = -0.61, R) >droYak2.chrX 7509953 94 - 21770863 AGCCCACGACUAAUUGUGUUCCC-ACAAACAUU--UUGCAGCCAUGGUGCGUAUGAACGUAUUGGCCGAUGCCCUGAAGUGCAUCAACAACGCCGAG ......((.....(((((....)-)))).....--..((.((((..((((((....)))))))))).(((((.(....).)))))......)))).. ( -22.70, z-score = -0.26, R) >droEre2.scaffold_4690 5106075 93 + 18748788 UACUCUCGACUAAUUGUGUUCCC--CAAACAUU--UUGCAGCCAUGGUGCGUAUGAACGUAUUGGCCGAUGCCCUGAAGUGCAUCAACAACGCCGAG ....((((.......(((((...--..))))).--..((.((((..((((((....)))))))))).(((((.(....).)))))......)))))) ( -23.40, z-score = -1.16, R) >droAna3.scaffold_13335 2408799 84 + 3335858 --------ACUAAUCUCGAUUCUCUUUC-----UAUUGCAGCCAUGGUGCGUAUGAACGUAUUGGCCGAUGCCCUUAAGUGCAUCAACAACGCGGAG --------......(((((.......))-----...(((.((((..((((((....)))))))))).(((((.(....).)))))......)))))) ( -20.50, z-score = -0.72, R) >dp4.chrXL_group1a 3133518 96 + 9151740 -GCCGUGAACUAAUUUGUACUCUCGUUUUCACAUUUUGCAGCCAUGGUGCGUAUGAACGUAUUGGCCGAUGCCCUUAAAUGCAUCAACAACGCCGAG -...(((((..(((..(....)..)))))))).....((.((((..((((((....)))))))))).(((((........)))))......)).... ( -21.00, z-score = -0.06, R) >droPer1.super_25 1354769 96 - 1448063 -GCCGUGAACUAAUUUGUACUCUCGUUUUCACAUUUUGCAGCCAUGGUGCGUAUGAACGUAUUGGCCGAUGCCCUUAAAUGCAUCAACAACGCCGAG -...(((((..(((..(....)..)))))))).....((.((((..((((((....)))))))))).(((((........)))))......)).... ( -21.00, z-score = -0.06, R) >droWil1.scaffold_181150 1342565 94 - 4952429 ---UUCUUAUACCUGUGUGUUUUUUUUUAUUUCAUUUGCAGCCAUGGUGCGUAUGAACGUAUUGGCCGAUGCCCUUAAAUGCAUCAACAAUGCCGAA ---...........................(((((.((((.(....))))).))))).((((((...(((((........)))))..)))))).... ( -19.30, z-score = -0.36, R) >droMoj3.scaffold_6328 1615656 92 + 4453435 -----UGUAUAAAACGGAAACUGAUUUGUUUUUAAUUGCAGCCAUGGUGCGUAUGAACGUAUUGGCCGAUGCCCUGAAGUGCAUUAACAACGCCGAG -----((((.((((((((......))))))))....))))((((..((((((....)))))))))).(((((.(....).)))))............ ( -20.00, z-score = -0.01, R) >droVir3.scaffold_13042 4348771 89 + 5191987 --------GUGAAAAAGAAACUAAUUUGUUUUUAAUUGCAGCCAUGGUGCGUAUGAACGUAUUGGCCGAUGCCCUGAAGUGCAUUAACAACGCAGAA --------.......((((((......))))))..((((.((((..((((((....)))))))))).(((((.(....).)))))......)))).. ( -21.50, z-score = -1.17, R) >droGri2.scaffold_15081 3046078 92 + 4274704 -----GGUGUGAGAAAUAUAUUAAUUUGUACCUAAUUGCAGCCAUGGUGCGUAUGAACGUAUUGGCCGAUGCCCUGAAGUGCAUCAACAAUGCCGAG -----((((..((...........))..)))).....(((((((..((((((....)))))))))).(((((.(....).))))).....))).... ( -23.30, z-score = -0.92, R) >apiMel3.Group3 8964379 74 + 12341916 ----------------------AAAUACAUUUCUUUU-UAGCUAUGGUGCGUAUGAAUGUUCUCAGUGAUGCCCUCAAAUCCAUUAACAAUGCUGAA ----------------------.............((-((((...((.((((.(((......)))...)))))).................)))))) ( -10.25, z-score = 0.17, R) >triCas2.ChLG9 3591047 97 - 15222296 GAGGAGCGAGUAUUUGUGGCCUAACCUCACUUUUGUUGCAGAGAUGGUGCGGAUGAACGUAUUGAGUGAUGCGCUCAAGUCCAUCAACAAUGCCGAA ......((.(((((((..((..((......))..))..))))(((((((((......))))(((((((...)))))))..))))).....))))).. ( -24.40, z-score = 0.36, R) >consensus __C__UGGACUAAUUGUGUUCCA_UUUAACAUU__UUGCAGCCAUGGUGCGUAUGAACGUAUUGGCCGAUGCCCUGAAGUGCAUCAACAACGCCGAG .....................................((.((((..((((((....)))))))))).(((((........)))))......)).... (-12.97 = -13.65 + 0.68)

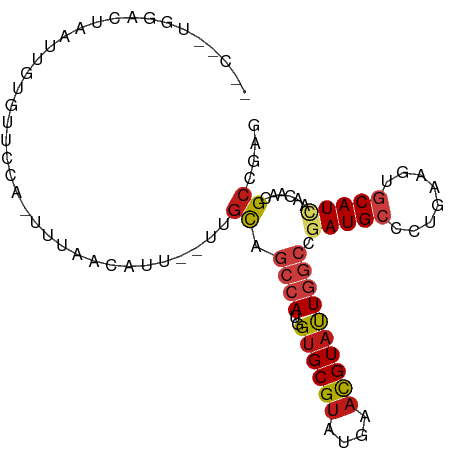

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:39:49 2011