
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,304,663 – 11,304,777 |
| Length | 114 |
| Max. P | 0.798722 |

| Location | 11,304,663 – 11,304,777 |
|---|---|
| Length | 114 |
| Sequences | 10 |
| Columns | 126 |
| Reading direction | forward |
| Mean pairwise identity | 79.91 |
| Shannon entropy | 0.41370 |
| G+C content | 0.45116 |
| Mean single sequence MFE | -29.04 |
| Consensus MFE | -20.11 |
| Energy contribution | -20.31 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.798722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
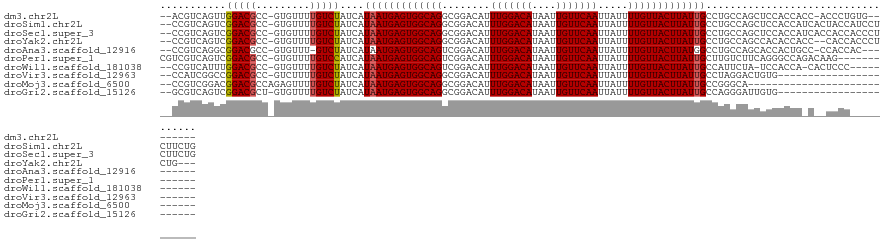
>dm3.chr2L 11304663 114 + 23011544 --ACGUCAGUUGGACGCC-GUGUUUUGUCUAUCAUAAUGAGUGGCAGGCGGACAUUUGGACAUAAUUGUUCAAUUAUUUUGUUACUUAUUGCCUGCCAGCUCCACCACC-ACCCUGUG-------- --....(((.((((((..-......)))))).......((((((((((((((((.(((((((....)))))))......))))......))))))))).))).......-...)))..-------- ( -31.10, z-score = -1.30, R) >droSim1.chr2L 11108393 123 + 22036055 --CCGUCAGUCGGACGCC-GUGUUUUGUCUAUCAUAAUGAGUGGCAGGCGGACAUUUGGACAUAAUUGUUCAAUUAUUUUGUUACUUAUUGCCUGCCAGCUCCACCAUCACUACCAUCCUCUUCUG --..(..(((.(((((..-......)))))........((((((((((((((((.(((((((....)))))))......))))......))))))))).))).......)))..)........... ( -29.80, z-score = -1.26, R) >droSec1.super_3 6701646 123 + 7220098 --CCGUCAGUCGGACGCC-GUGUUUUGUCUAUCAUAAUGAGUGGCAGGCGGACAUUUGGACAUAAUUGUUCAAUUAUUUUGUUACUUAUUGCCUGCCAGCUCCACCAUCACCACCACCCUCUUCUG --....(((..(((((..-......)))))........((((((((((((((((.(((((((....)))))))......))))......))))))))).))).....................))) ( -29.50, z-score = -1.07, R) >droYak2.chr2L 7701564 118 + 22324452 --CCGUCAGUCGGACGCC-GUGUUUUGUCUAUCAUAAUGAGUGGCAGGCGGACAUUUGGACAUAAUUGUUCAAUUAUUUUGUUACUUAUUGCCUGCCAGCCACACCACC--CACCACCCUCUG--- --.((((.....))))..-((((........((.....)).(((((((((((((.(((((((....)))))))......))))......)))))))))...))))....--............--- ( -28.30, z-score = -0.62, R) >droAna3.scaffold_12916 12025454 112 - 16180835 --CCGUCAGGCGGACGCC-GUGUUU-GUCUAUCAUAAUGAGUGGCAGUCGGACAUUUGGACAUAAUUGUUCAAUUAUUUUGUUACUUAUGGCCUGCCAGCACCACUGCC-CCACCAC--------- --......(((((..(((-((((((-(.((..(((.....)))..)).)))))))(((((((....)))))))................)))))))).(((....))).-.......--------- ( -30.70, z-score = -0.65, R) >droPer1.super_1 8189716 112 + 10282868 CGUCGUCAGUCGGACGCC-GUGUUUUGUCCAUCAUAAUGAGUGGCAGUCGGACAUUUGGACAUAAUUGUUCAAUUAUUUUGUUACUUAUUGCUUGUCUUCAGGGCCAGACAAG------------- ((.((((.....)))).)-)(((((.((((....(((((((((((((........(((((((....))))))).....)))))))))))))..((....)))))).)))))..------------- ( -32.02, z-score = -2.12, R) >droWil1.scaffold_181038 216641 110 - 637489 --CCGUCAUUUGGACGCC-GUGUUUUGUCUAUCAUAAUGAGUGGCAGUCGGACAUUUGGACAUAAUUGUUCAAUUAUUUUGUUACUUAUUGCCAUUCUA-UCCACCA-CACUCCC----------- --.((((.....))))..-((((..((..((.......((((((((((..((((.(((((((....)))))))......))))....))))))))))))-..))..)-)))....----------- ( -27.21, z-score = -2.18, R) >droVir3.scaffold_12963 17994035 100 - 20206255 --CCAUCGGCCGGACGCC-GUCUUUUGUCUAUCAUAAUGAGUGGCAGGCGGACAUUUGGACAUAAUUGUUCAAUUAUUUUGUUACUUAUUGCCUAGGACUGUG----------------------- --....((((.....)))-)......((((....((((((((((((((.......(((((((....)))))))....))))))))))))))....))))....----------------------- ( -26.90, z-score = -1.29, R) >droMoj3.scaffold_6500 4937343 96 - 32352404 --CCGUCGGACGGACGCCAGAGUUUUGUCUAUCAUAAUGAGUGGCAGGCGGACAUUUGGACAUAAUUGUUCAAUUAUUUUGUUACUUAUUGCCGGGCA---------------------------- --(((..((((((((......).)))))))....((((((((((((((.......(((((((....)))))))....)))))))))))))).)))...---------------------------- ( -26.80, z-score = -1.41, R) >droGri2.scaffold_15126 7109080 100 - 8399593 --GCGUCAGUCGGACGCU-GUGUUUUGUCUAUCAUAAUGAGUGGCAGGCGGACAUUUGGACAUAAUUGUUCAAUUAUUUUGUUACUUAUUGCCAGGGAUUGUG----------------------- --(((((.....))))).-.......((((....((((((((((((((.......(((((((....)))))))....))))))))))))))....))))....----------------------- ( -28.10, z-score = -1.70, R) >consensus __CCGUCAGUCGGACGCC_GUGUUUUGUCUAUCAUAAUGAGUGGCAGGCGGACAUUUGGACAUAAUUGUUCAAUUAUUUUGUUACUUAUUGCCUGCCAGCUCCACCA_C__CACC___________ ...((((.....))))..................(((((((((((((........(((((((....))))))).....)))))))))))))................................... (-20.11 = -20.31 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:02 2011