| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,225,752 – 13,225,855 |
| Length | 103 |
| Max. P | 0.917003 |

| Location | 13,225,752 – 13,225,855 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 60.96 |
| Shannon entropy | 0.72089 |
| G+C content | 0.48221 |
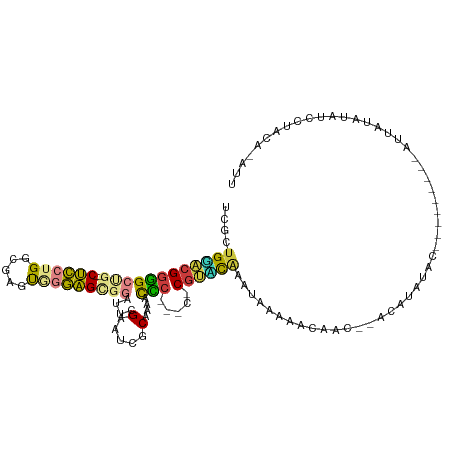
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -11.51 |
| Energy contribution | -11.09 |
| Covariance contribution | -0.42 |
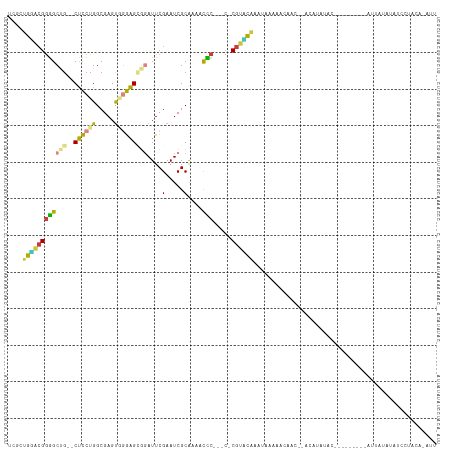
| Combinations/Pair | 1.56 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.917003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
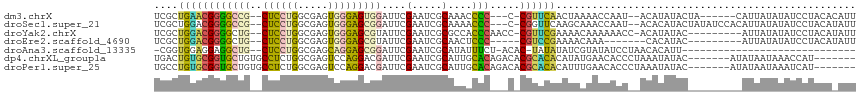
>dm3.chrX 13225752 103 + 22422827 UCGCUGAACGGGGCCG--CUCCUGGCGAGUGGGAGUGGAUUCGAAUCGCAAACCCC---C-CGUUCAACUAAAACCAAU--ACAUAUACUA------CAUUAUAUAUCCUACACAUU ....((((((((((((--(((((.......))))))))....(.....).....))---)-))))))............--..(((((...------...)))))............ ( -27.40, z-score = -1.78, R) >droSec1.super_21 980926 109 + 1102487 UCGCUGGACGGGGCCG--CUCCUGGCGAGUGGGAGCGGAUUCGAAUCGCAAAACCC---C-CGGUUCAAGCAAACCAAU--ACACAUACUAUAUCCACAUUAUAUAUCCUACAUAUU ..((((((((((((((--(((((.......))))))))....(.....).....))---)-).)))).)))........--.................................... ( -28.20, z-score = -0.63, R) >droYak2.chrX 7504293 104 + 21770863 UCGCUGGACGGGGCUG--CUCCUGGCGAGUGGGAGCGUAUUCGAAUCGCGCCACCCAACC-CGUUCGAAAACAAAAAACC-ACAUAUAC---------AUUAUAUAUCCUACAUAUU ....(((((((((((.--.....)))(.((((..(((.........))).)))).)..))-)))))).............-........---------................... ( -24.90, z-score = -0.27, R) >droEre2.scaffold_4690 5101808 94 - 18748788 UCGCUGGACGGGGCUG--CUCCUGGCGAGUGGGAGCGUAUUCGAAUCGCAACUCCC-----CGUCCGAAAACAAA-------CACAUAC---------AUUAUAUAUCCUACAUAUU ....((((((((((..--(((.....)))..)..(((.........)))....)))-----))))))........-------.......---------................... ( -26.40, z-score = -1.32, R) >droAna3.scaffold_13335 2403888 83 - 3335858 -CGGUGGAGGAGGCUG--CUCCUGGCGAGCAGGAGCGGAUUCGAAUCGCAUAUUUCU-ACAC-UAUAUAUCGUAUAUCCUAACACAUU----------------------------- -..(((.((((..(((--((((((.....)))))))))....(((........))).-....-.............))))..)))...----------------------------- ( -23.60, z-score = -1.31, R) >dp4.chrXL_group1a 3129518 103 - 9151740 UGACUGUGCGGUGCUGUGCCUCUGGCGAGUCCAGGACGAUUCGAAUCGCAUUGCACAGACACGCACACAUAUGAACACCCUAAAUAUAC-------AUAUAAUAAACCAU------- ....((((((((.((((((.(((((.....))))).((((....))))....)))))))).)))))).(((((...............)-------))))..........------- ( -26.76, z-score = -1.68, R) >droPer1.super_25 1350742 103 + 1448063 UGCCUGUGCGGUGCUGUGCCUCUGGCGAGUCCAGGACGAUUCGAAUCGCAUUGCACAGACACGCACACAUUUGAACACCCUAAAUAUAC-------AUAUAAUAAAUCAU------- ((((((((((((((...(((...)))(((((......))))).....)))))))))))....)))........................-------..............------- ( -26.50, z-score = -1.24, R) >consensus UCGCUGGACGGGGCUG__CUCCUGGCGAGUGGGAGCGGAUUCGAAUCGCAAAACCC___C_CGUACAAAUAAAAACAAC__ACAUAUAC_________AUUAUAUAUCCUACA_AUU ....(((((((((.....))))..((((...(((.....)))...)))).............))))).................................................. (-11.51 = -11.09 + -0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:39:49 2011