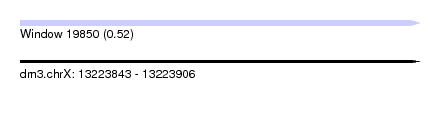
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,223,843 – 13,223,906 |
| Length | 63 |
| Max. P | 0.517347 |

| Location | 13,223,843 – 13,223,906 |
|---|---|
| Length | 63 |
| Sequences | 9 |
| Columns | 75 |
| Reading direction | forward |
| Mean pairwise identity | 62.88 |
| Shannon entropy | 0.70307 |
| G+C content | 0.42020 |
| Mean single sequence MFE | -12.42 |
| Consensus MFE | -4.58 |
| Energy contribution | -4.48 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.517347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13223843 63 + 22422827 ----------UUCGCUUAAUUU-CAAUCCAUUCGCGAAUUUCAACUCGUGCCGAAAGGUUU-GUUUGUGCGAAAG ----------(((((.(((...-(((.......((((........))))(((....)))))-).))).))))).. ( -14.80, z-score = -1.53, R) >droAna3.scaffold_13335 2402055 57 - 3335858 ------------CACCUAUUAUUCGCAGUAUUUCGUUUUUGAUUCGCGUGCCGAAAGGUUG------UGCUUAAA ------------......(((..((((((...(((....)))...))..(((....)))))------))..))). ( -10.10, z-score = -0.35, R) >droEre2.scaffold_4690 5099953 60 - 18748788 ----------UACCACUAAAGUUCAAUCCAUUCGCGAAUUUCAACUCGUGCCGAAAGGUUU-G----UGCGAAAG ----------....................(((((((........))..(((....)))..-.----.))))).. ( -11.30, z-score = -1.04, R) >droYak2.chrX 7502354 60 + 21770863 ----------UACCACUAAAUUUCAAUCCAUUCGCGAAUUUCAACUCGUGCCGAAAGGUUU-G----UGCGAAAG ----------....................(((((((........))..(((....)))..-.----.))))).. ( -11.30, z-score = -1.31, R) >droSec1.super_21 979137 64 + 1102487 ----------UACACUUAAUUUUCAAUUCAUUCGCGAAUUUCAACUCGUGCCGAAAGGUUU-GUUUGUGCGAAAG ----------....................((((((.....(((.....(((....)))))-)....)))))).. ( -12.70, z-score = -1.10, R) >droSim1.chrX 10114858 64 + 17042790 ----------UACACUUAAUUUUCAAUUCAUUCGCGAAUUUCAACUCGUGCCGAGAGGUUU-GUUUGUGCGAAAG ----------....................((((((.....(((.....(((....)))))-)....)))))).. ( -11.70, z-score = -0.36, R) >droPer1.super_25 1349341 75 + 1448063 UACUGUAUACCAUAGCUACAUCUUCCUCCAUUCGCGAUUUCGUUACCGUGCCGGAAGGUUGUGCUUUUUCUCUGG .........(((.(((.(((((((((..(((..(((....)))....)))..)))))).)))))).......))) ( -15.10, z-score = -1.01, R) >droWil1.scaffold_181096 5845268 60 - 12416693 ----------UUCGCAUAAAA---AAAUCAUUUUCAAAUUUCCAC-CAAGUCGAAAGGUUG-GCGAGUGCGGAAG ----------(((((((((((---......))))........(.(-(((.((....)))))-).).))))))).. ( -12.50, z-score = -1.54, R) >dp4.chrXL_group1a 3128127 60 - 9151740 ---------------CUAAAUCUUCCUCCAUUCGCGAUUUCGUUACCGUGCCGGAAGGUUGUGCUUUUUCUCUGG ---------------...((((((((..(((..(((....)))....)))..))))))))............... ( -12.30, z-score = -1.20, R) >consensus __________UACACCUAAAUUUCAAUCCAUUCGCGAAUUUCAACUCGUGCCGAAAGGUUU_G_UUGUGCGAAAG .................................................(((....)))................ ( -4.58 = -4.48 + -0.10)

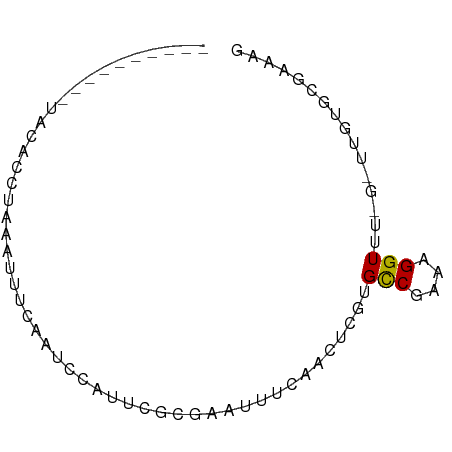
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:39:48 2011