
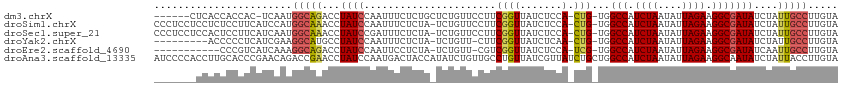
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,207,311 – 13,207,451 |
| Length | 140 |
| Max. P | 0.616416 |

| Location | 13,207,311 – 13,207,421 |
|---|---|
| Length | 110 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.98 |
| Shannon entropy | 0.48373 |
| G+C content | 0.41164 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -10.39 |
| Energy contribution | -10.96 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.616416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13207311 110 + 22422827 ----UGCCAAUUGCUGAUAAGG-CGCAACAAGCGCACUUGUACAAGGCAAUAGAUAUCGCCUUCUAAUAUUAGAUGGCCAC--AGUGGAGAUAACCGAAGGAACAGAGCAGAGAAAU--- ----((((.((((((.((((((-(((.....)))).)))))....)))))).....(((((.((((....)))).)))..(--..(((......)))..)))...).))).......--- ( -28.90, z-score = -2.14, R) >droSim1.chrX 10097213 109 + 17042790 ----UGCCAAUUGCUGAUAAGG-CGCAACAAGCGCACUUGUACAAGGCAAUAGAUAUCGCCUUCUAAUAUUAGAUGGCCAC--AGUGGAGAUAACCGAAGGAACAGA-UAGAGAAAU--- ----.....((((((.((((((-(((.....)))).)))))....))))))...(((((((.((((....)))).)))..(--..(((......)))..).....))-)).......--- ( -28.90, z-score = -2.64, R) >droSec1.super_21 962668 109 + 1102487 ----UGCCAAUUGCUGAUAAGG-CGCAACAAGCGCACUUGUACAAGGCAAUAGAUAUCGCCUUCUAAUAUUAGAUGGCCAC--AGUGGAGAUAACCGAAGGAACAGA-UAGAGAAAU--- ----.....((((((.((((((-(((.....)))).)))))....))))))...(((((((.((((....)))).)))..(--..(((......)))..).....))-)).......--- ( -28.90, z-score = -2.64, R) >droYak2.chrX 7485858 108 + 21770863 ----UGGCAAUUGCUGAUAAGG-CGAAACAAGUGCACUUGUACAAGGCAAUAGAUAUCGCCUUCUAAUAUUAGAUGGCCAC--AGUUGAGAUAACCGAAG-AACAGA-UAGAGAAAU--- ----((((.....(((((((((-(((......(((.(((....)))))).......)))))).....))))))...)))).--.................-......-.........--- ( -23.12, z-score = -1.48, R) >droEre2.scaffold_4690 5082838 108 - 18748788 ----UGGCAAUUGCUGAUAAGG-UGAAACAAGAGCACUUGUACAAGGCAAUUGAUAUCGCCUUCUAAUAUUAGAUGGCCAC--GAUGGAGAUAACCGACG-AACAGA-UAGAGGAAU--- ----...((((((((.((((((-(.........)).)))))....)))))))).(((((((.((((....)))).)))..(--(.(((......))).))-....))-)).......--- ( -26.40, z-score = -2.56, R) >droAna3.scaffold_13335 2376734 117 - 3335858 UUUGGGCCAAUUGGCGAUAAGGCUUAGACAAGAGCACUUGUACAAGGUAAUAGAUAUUGCCUUCUAAUAUUAGAUGGCCAGCAGAUAACGAUAACAGGC--AACAGA-UAUGGUAGUCAU ....(((((.(((...(((((((((......)))).))))).)))((((((....)))))).((((....)))))))))....(((.((.(((.(.(..--..).).-))).)).))).. ( -31.20, z-score = -2.40, R) >droPer1.super_25 1333699 84 + 1448063 ----AGGCAACAGUUGAUAAGGGGUAAAUAA------UUGUUU--GGAAAUAGAUAUCGCUCUCC--CCAGAGAUAACCGA--UGAAAAGAUAAAGAAAU-------------------- ----.(....).........((((.......------.(((((--(....)))))).......))--))............--.................-------------------- ( -10.86, z-score = 0.62, R) >consensus ____UGCCAAUUGCUGAUAAGG_CGAAACAAGCGCACUUGUACAAGGCAAUAGAUAUCGCCUUCUAAUAUUAGAUGGCCAC__AGUGGAGAUAACCGAAG_AACAGA_UAGAGAAAU___ .........((((((.(((((...............)))))....)))))).......(((.((((....)))).))).......................................... (-10.39 = -10.96 + 0.57)



| Location | 13,207,346 – 13,207,451 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.44 |
| Shannon entropy | 0.43086 |
| G+C content | 0.43842 |
| Mean single sequence MFE | -19.10 |
| Consensus MFE | -12.08 |
| Energy contribution | -12.80 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
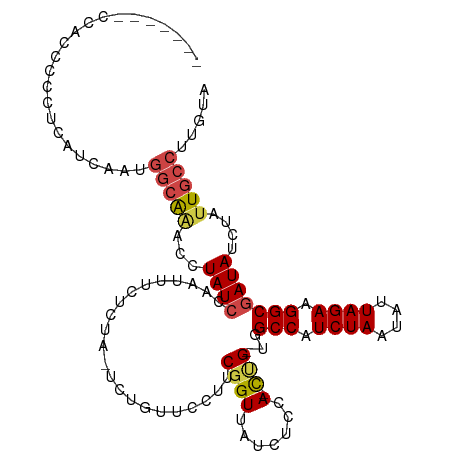
>dm3.chrX 13207346 105 - 22422827 ------CUCACCACCAC-UCAAUGGCAGACCUAUCCAAUUUCUCUGCUCUGUUCCUUCGGUUAUCUCCA-CUG-UGGCCAUCUAAUAUUAGAAGGCGAUAUCUAUUGCCUUGUA ------.......((((-.....((((((.............))))))..........((......)).-..)-)))...........((.((((((((....)))))))).)) ( -21.62, z-score = -1.47, R) >droSim1.chrX 10097248 111 - 17042790 CCCUCCUCCUCUCCUUCAUCCAUGGCAAACCUAUCCAAUUUCUCUA-UCUGUUCCUUCGGUUAUCUCCA-CUG-UGGCCAUCUAAUAUUAGAAGGCGAUAUCUAUUGCCUUGUA ....................((.(((((..................-...........((......)).-.((-(.(((.((((....)))).))).)))....))))).)).. ( -18.70, z-score = -1.84, R) >droSec1.super_21 962703 111 - 1102487 CCCUCCUCCACUCCUUCAUCAAUGGCAAACCUAUCCGAUUUCUCUA-UCUGUUCCUUCGGUUAUCUCCA-CUG-UGGCCAUCUAAUAUUAGAAGGCGAUAUCUAUUGCCUUGUA ...................(((.(((((......((((.....(..-...).....)))).........-.((-(.(((.((((....)))).))).)))....)))))))).. ( -20.80, z-score = -2.03, R) >droYak2.chrX 7485893 101 - 21770863 ---------ACCCCCUCAUCGAAGGCAUGCCUAUCCAAUUUCUCUA-UCUGUU-CUUCGGUUAUCUCAA-CUG-UGGCCAUCUAAUAUUAGAAGGCGAUAUCUAUUGCCUUGUA ---------............((((((....((((...........-......-...(((((.....))-)))-..(((.((((....)))).))))))).....))))))... ( -19.90, z-score = -1.07, R) >droEre2.scaffold_4690 5082873 99 + 18748788 -----------CCCGUCAUCAAAGGCAGACCUAUCCAAUUCCUCUA-UCUGUU-CGUCGGUUAUCUCCA-UCG-UGGCCAUCUAAUAUUAGAAGGCGAUAUCAAUUGCCUUGUA -----------..........(((((((...((((...........-......-((..((......)).-.))-..(((.((((....)))).)))))))....)))))))... ( -20.40, z-score = -1.25, R) >droAna3.scaffold_13335 2376774 114 + 3335858 AUCCCCACCUUGCACCCGAACAGACCGAACCUAUCCAAUGACUACCAUAUCUGUUGCCUGUUAUCGUUAUCUGCUGGCCAUCUAAUAUUAGAAGGCAAUAUCUAUUACCUUGUA ..........((((....((.(((.............(((.....)))...((((((((.(((..((((..((.....))..))))..))).))))))))))).))....)))) ( -13.20, z-score = 1.02, R) >consensus _______CCACCCCCUCAUCAAUGGCAAACCUAUCCAAUUUCUCUA_UCUGUUCCUUCGGUUAUCUCCA_CUG_UGGCCAUCUAAUAUUAGAAGGCGAUAUCUAUUGCCUUGUA .......................(((((...((((.......................((......))........(((.((((....)))).)))))))....)))))..... (-12.08 = -12.80 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:39:47 2011