
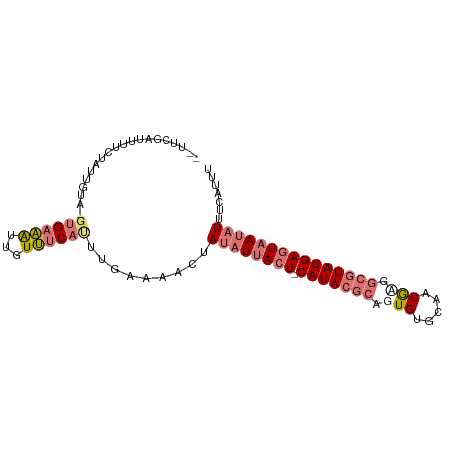
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,195,845 – 13,195,936 |
| Length | 91 |
| Max. P | 0.999501 |

| Location | 13,195,845 – 13,195,936 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 71.15 |
| Shannon entropy | 0.46522 |
| G+C content | 0.31293 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -12.45 |
| Energy contribution | -14.45 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.998535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
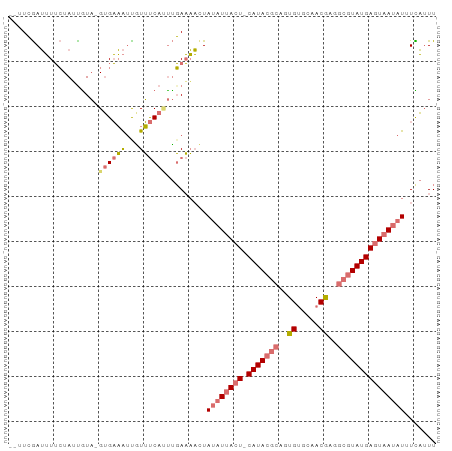
>dm3.chrX 13195845 91 + 22422827 --UUCGAUUUUCUAUUGUA-GUGAAAUUGUUUCAUUUGAAAACUAUAUUACU-CAUACGCAAUGUGCAACGAGGCGUAUGAGUAAUAUUUCAUUU --................(-((((((...((((....))))....(((((((-(((((((..((.....))..))))))))))))))))))))). ( -25.70, z-score = -4.02, R) >droSec1.super_21 951392 91 + 1102487 --UUCGAUUUUCUAUUGUA-GUGAAAUCGUUUCAUUUGAAAACUAUAUUACU-CAUACGCACUGUGCAACGAGGCGUAUGAGUAAUAUGUCAUUU --..((((((((((...))-).)))))))((((....))))..(((((((((-(((((((..((.....))..))))))))))))))))...... ( -28.40, z-score = -4.45, R) >droYak2.chrX 7473230 91 + 21770863 --GUCGAUUUUCUAGCAUA-GUGAAAUUAUUUCACUUGAAAAGUAUAUUACU-CAUACGCAGUGGGCAACGCGGCGUAUGAGUAAUAUUUUAGUU --.....(((((......(-(((((.....)))))).)))))..((((((((-(((((((.(((.....))).)))))))))))))))....... ( -31.10, z-score = -5.36, R) >droAna3.scaffold_12916 7698174 95 + 16180835 CCUUAAAAAUAGUAAUGUUUGAGUUGAUGUACCAGCCUAUUUUUAGUUAAAUACAUAAACUGUGUGACACAUGUCGUAUGAGUAAUAUUUUUUUU ....(((((((..........((((.(((((.....(((....))).....))))).))))(((((((....)))))))......)))))))... ( -12.00, z-score = 0.53, R) >consensus __UUCGAUUUUCUAUUGUA_GUGAAAUUGUUUCAUUUGAAAACUAUAUUACU_CAUACGCAGUGUGCAACGAGGCGUAUGAGUAAUAUUUCAUUU ............................................((((((((.(((((((.(((.....))).)))))))))))))))....... (-12.45 = -14.45 + 2.00)

| Location | 13,195,845 – 13,195,936 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 71.15 |
| Shannon entropy | 0.46522 |
| G+C content | 0.31293 |
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -9.69 |
| Energy contribution | -11.38 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -4.19 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.95 |
| SVM RNA-class probability | 0.999501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
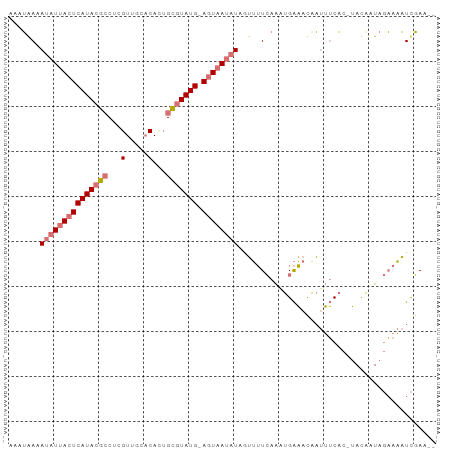
>dm3.chrX 13195845 91 - 22422827 AAAUGAAAUAUUACUCAUACGCCUCGUUGCACAUUGCGUAUG-AGUAAUAUAGUUUUCAAAUGAAACAAUUUCAC-UACAAUAGAAAAUCGAA-- ...(((((((((((((((((((...(.....)...)))))))-)))))))).(((((.....)))))...)))).-.................-- ( -22.90, z-score = -4.41, R) >droSec1.super_21 951392 91 - 1102487 AAAUGACAUAUUACUCAUACGCCUCGUUGCACAGUGCGUAUG-AGUAAUAUAGUUUUCAAAUGAAACGAUUUCAC-UACAAUAGAAAAUCGAA-- ....((((((((((((((((((((.(.....))).)))))))-)))))))).)))...........((((((..(-((...))).))))))..-- ( -26.30, z-score = -4.74, R) >droYak2.chrX 7473230 91 - 21770863 AACUAAAAUAUUACUCAUACGCCGCGUUGCCCACUGCGUAUG-AGUAAUAUACUUUUCAAGUGAAAUAAUUUCAC-UAUGCUAGAAAAUCGAC-- ..(((..(((((((((((((((.(.(.....).).)))))))-))))))))........((((((.....)))))-)....))).........-- ( -26.50, z-score = -5.75, R) >droAna3.scaffold_12916 7698174 95 - 16180835 AAAAAAAAUAUUACUCAUACGACAUGUGUCACACAGUUUAUGUAUUUAACUAAAAAUAGGCUGGUACAUCAACUCAAACAUUACUAUUUUUAAGG ...(((((((..........(((....)))....((((.(((((((...(((....)))...))))))).))))..........))))))).... ( -14.10, z-score = -1.87, R) >consensus AAAUAAAAUAUUACUCAUACGCCUCGUUGCACACUGCGUAUG_AGUAAUAUAGUUUUCAAAUGAAACAAUUUCAC_UACAAUAGAAAAUCGAA__ .......(((((((((((((((.(.(.....).).))))))).))))))))............................................ ( -9.69 = -11.38 + 1.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:39:45 2011