| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,139,707 – 13,139,802 |
| Length | 95 |
| Max. P | 0.992375 |

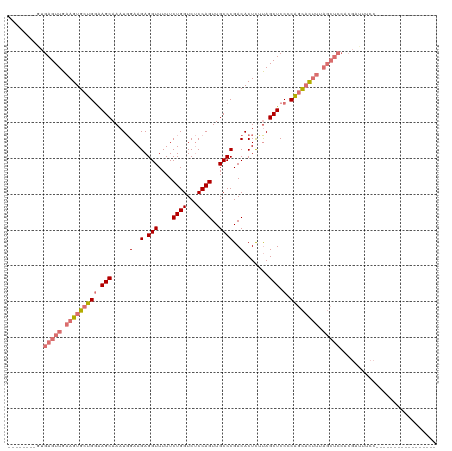
| Location | 13,139,707 – 13,139,802 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.78 |
| Shannon entropy | 0.41563 |
| G+C content | 0.50359 |
| Mean single sequence MFE | -32.85 |
| Consensus MFE | -21.46 |
| Energy contribution | -23.90 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.992375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13139707 95 + 22422827 --------AUGAGUUGAAGUGCUGGAAGCAAAUGGAAGAGGUUUUUCUGGUCUCCAGUUGCCCGACACUCACAGCUCCCCAGCACUUUAGCUCCACGAUUUCC----------------- --------..(((((((((((((((.(((........(.(((....((((...))))..))))(.....)...)))..)))))))))))))))..........----------------- ( -34.40, z-score = -3.08, R) >droSim1.chrX 10061783 79 + 17042790 --------GUGAGUUGAAGUGCUGCAAGCAAAUGGAAGAGGUUUUUCUGGUCUCCAGUUGCCCGACACUUUUAGCUCCACGAUUUCC--------------------------------- --------(((((((((((((.((...((((.((((...((.....))....)))).)))).)).))).))))))).))).......--------------------------------- ( -20.70, z-score = -0.58, R) >droSec1.super_21 883730 95 + 1102487 --------GUGAGUUGAAGUGCUGGAAGCAAAUGGAAGAGGUUUUUCUGGUCUCCAGUUGCCCGACACUUUUAGCUCCGCAGCACUUUAGCUCCACGAUUUCC----------------- --------(((((((((((((((((.((((((..(..(.(((....((((...))))..))))..)...))).))).).))))))))))))).))).......----------------- ( -34.80, z-score = -3.02, R) >droYak2.chrX 7411766 120 + 21770863 GUGAGUGAGUGAGUUGAAGUGCUGGGAGCAAAUCGAAGAGGUUUUUCUGGUCCCCAGUUGCCCGACACUCGUAGCUCCCCAGCACUUAAGCUCCCCGCUUUCCGAACUCCAAUUCCCAUU ..(((((.(.(((((.(((((((((((((....(((.(.(((....((((...))))..)))).....)))..))).)))))))))).)))))).))))).................... ( -41.50, z-score = -3.43, R) >consensus ________GUGAGUUGAAGUGCUGGAAGCAAAUGGAAGAGGUUUUUCUGGUCUCCAGUUGCCCGACACUCUUAGCUCCCCAGCACUUUAGCUCCACGAUUUCC_________________ ..........(((((((((((((((.(((.....(..(.(((....((((...))))..))))..).......))).).))))))))))))))........................... (-21.46 = -23.90 + 2.44)


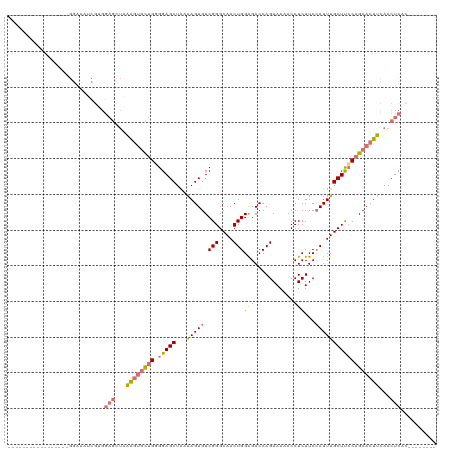
| Location | 13,139,707 – 13,139,802 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.78 |
| Shannon entropy | 0.41563 |
| G+C content | 0.50359 |
| Mean single sequence MFE | -29.65 |
| Consensus MFE | -20.84 |
| Energy contribution | -21.90 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.591894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13139707 95 - 22422827 -----------------GGAAAUCGUGGAGCUAAAGUGCUGGGGAGCUGUGAGUGUCGGGCAACUGGAGACCAGAAAAACCUCUUCCAUUUGCUUCCAGCACUUCAACUCAU-------- -----------------..........(((...((((((((((.(((.(((.....(((....)))(((...........)))...)))..)))))))))))))...)))..-------- ( -29.30, z-score = -0.69, R) >droSim1.chrX 10061783 79 - 17042790 ---------------------------------GGAAAUCGUGGAGCUAAAAGUGUCGGGCAACUGGAGACCAGAAAAACCUCUUCCAUUUGCUUGCAGCACUUCAACUCAC-------- ---------------------------------..........(((....(((((((((((((.((((....(((......))))))).)))))))..))))))...)))..-------- ( -20.80, z-score = -1.10, R) >droSec1.super_21 883730 95 - 1102487 -----------------GGAAAUCGUGGAGCUAAAGUGCUGCGGAGCUAAAAGUGUCGGGCAACUGGAGACCAGAAAAACCUCUUCCAUUUGCUUCCAGCACUUCAACUCAC-------- -----------------..........(((...((((((((.(((((...(((((.(((....)))(((...........)))...))))))))))))))))))...)))..-------- ( -28.60, z-score = -1.45, R) >droYak2.chrX 7411766 120 - 21770863 AAUGGGAAUUGGAGUUCGGAAAGCGGGGAGCUUAAGUGCUGGGGAGCUACGAGUGUCGGGCAACUGGGGACCAGAAAAACCUCUUCGAUUUGCUCCCAGCACUUCAACUCACUCACUCAC ..((((.....(((((.(((((((.....))))..((((((((.(((..((((....((....))((((..........))))))))....))))))))))))))))))).....)))). ( -39.90, z-score = -0.95, R) >consensus _________________GGAAAUCGUGGAGCUAAAGUGCUGGGGAGCUAAAAGUGUCGGGCAACUGGAGACCAGAAAAACCUCUUCCAUUUGCUUCCAGCACUUCAACUCAC________ ...........................(((...((((((((.(((((...(((((.(((....)))(((...........)))...))))))))))))))))))...))).......... (-20.84 = -21.90 + 1.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:39:37 2011