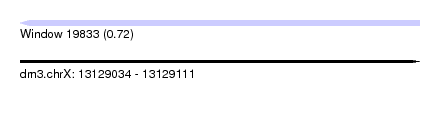
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,129,034 – 13,129,111 |
| Length | 77 |
| Max. P | 0.718966 |

| Location | 13,129,034 – 13,129,111 |
|---|---|
| Length | 77 |
| Sequences | 8 |
| Columns | 86 |
| Reading direction | reverse |
| Mean pairwise identity | 72.67 |
| Shannon entropy | 0.52881 |
| G+C content | 0.35477 |
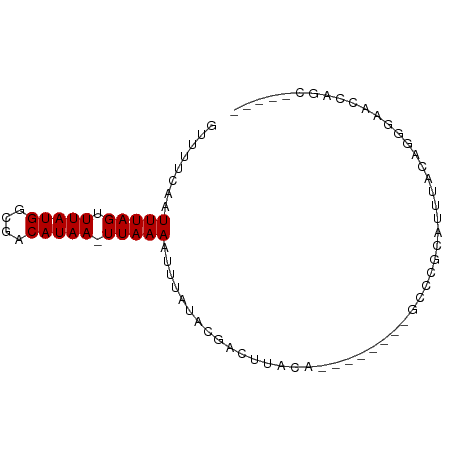
| Mean single sequence MFE | -12.29 |
| Consensus MFE | -7.00 |
| Energy contribution | -7.00 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.718966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
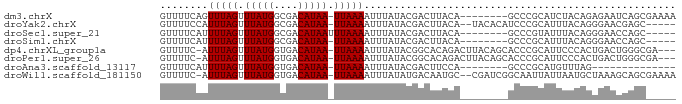
>dm3.chrX 13129034 77 - 22422827 GUUUUCAGUUUAGUUUAUGGCGACAUAA-UUAAAAUUUAUACGACUUACA--------GCCCGCAUCUACAGAGAAUCAGCGAAAA ((......(((((((.(((....)))))-)))))......))........--------...(((.(((....)))....))).... ( -9.30, z-score = -0.02, R) >droYak2.chrX 7399988 78 - 21770863 GUUUUCCAUUUAGUUUAUGGCGACAUAA-UUAAAAUUUAUACGACUUACA--UACACAUCCCGCAUUUACAGGGAACGAGC----- ........(((((((.(((....)))))-)))))................--......((((.........))))......----- ( -11.00, z-score = -1.39, R) >droSec1.super_21 874047 73 - 1102487 GUUUUCAUUUUAGUUUAUGGCGACAUAAUUUAAAAUUUAUACGACUUACA--------GCCCGUAUUUACAGGGAACCAGC----- (((..((((((((.(((((....))))).)))))))..(((((.......--------...))))).....)..)))....----- ( -14.10, z-score = -2.47, R) >droSim1.chrX 10019350 72 - 17042790 GUUUUCAUUUUAGUUUAUGGCGACAUAA-UUAAAAUUUAUACGACUUACA--------GCCCGCAUUUACAGGGAACCAGC----- ((....(((((((((.(((....)))))-)))))))....))........--------.(((.........))).......----- ( -11.60, z-score = -1.61, R) >dp4.chrXL_group1a 8298393 81 + 9151740 GUUUUC-AUUUAGUUUAUGGUGACAUAA-UUAAAAUUUAUACGGCACAGACUUACAGCACCCGCAUUCCCACUGACUGGGCGA--- ......-.(((((((.(((....)))))-))))).......((.............((....))...((((.....)))))).--- ( -13.00, z-score = -0.02, R) >droPer1.super_26 384126 81 + 1349181 GUUUUC-AUUUAGUUUAUGGUGACAUAA-UUAAAAUUUAUACGGCACAGACUUACAGCACCCGCAUUCCCACUGACUGGGCGA--- ......-.(((((((.(((....)))))-))))).......((.............((....))...((((.....)))))).--- ( -13.00, z-score = -0.02, R) >droAna3.scaffold_13117 4026116 63 - 5790199 GUUUUCAUUUUAGUUUAUGGUGACAUAA-UUAAAAUUUAUACGACUUCCA--------GCCCGCAUGUUUAG-------------- ((....(((((((((.(((....)))))-)))))))....))........--------..............-------------- ( -9.40, z-score = -1.57, R) >droWil1.scaffold_181150 1228584 82 + 4952429 GUUUUC-AUUUAGUUUAUGGUGACAUAA-UUAAAAUUUAUAUGACAAUGC--CGAUCGGCAAUUAUUAAUGCUAAAGCAGCGAAAA .(((((-.(((((((.(((....)))))-))))).............(((--(....))))........(((....)))..))))) ( -16.90, z-score = -1.44, R) >consensus GUUUUCAAUUUAGUUUAUGGCGACAUAA_UUAAAAUUUAUACGACUUACA________GCCCGCAUUUACAGGGAACCAGC_____ ........(((((.(((((....))))).))))).................................................... ( -7.00 = -7.00 + -0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:39:34 2011