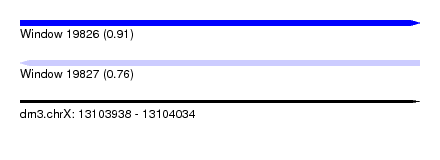
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,103,938 – 13,104,034 |
| Length | 96 |
| Max. P | 0.911634 |

| Location | 13,103,938 – 13,104,034 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 73.13 |
| Shannon entropy | 0.49464 |
| G+C content | 0.49949 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -19.14 |
| Energy contribution | -18.84 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
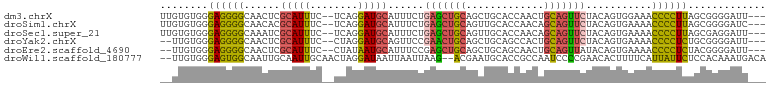
>dm3.chrX 13103938 96 + 22422827 UUGUGUGGGAGGGGCAACUCGCAUUUC--UCAGGAUGCAUUUCUGAGCUGCAGCUGCACCAACUGCAGUUCUACAGUGGAAACCCCUUAGCGGGGAUU--- ...((((((.(((....)))(((...(--((((((.....))))))).))).((((((.....)))))))))))).......((((.....))))...--- ( -34.80, z-score = -1.24, R) >droSim1.chrX 9993310 96 + 17042790 UUGUGUGGGAGGGGCAACACGCAUUUC--UCAGGAUGCAUUUCUGAGCUGCAGUUGCACCAACAGCAGUUCUACAGUGAAAACCCCUUAGCGGGGAUC--- ((.(((..((((((...((((((((..--....)))))......(((((((.((((...)))).)))))))....)))....)))))).))).))...--- ( -33.20, z-score = -1.56, R) >droSec1.super_21 849369 96 + 1102487 UUGUGUGGGAGGGGCAAAUCGCAUUUC--UCAGGAUGCAUUUCUGAGCUGCAGUUGCACCAACAGCAGUUCUACAGUGAAAACCCCUUAGCGAGGAUU--- ((((....((((((....((((....(--((((((.....)))))))((((.((((...)))).)))).......))))...)))))).)))).....--- ( -30.80, z-score = -1.17, R) >droYak2.chrX 7375076 94 + 21770863 --UUGUGGGAGGGGCAACUCGCAUUUC--CUAGGAUGCAGUUCCGAACUGCAGCUGCAGCCACUGCAGUUCUACAGUGAAAACCCCUCUGCGGGGAUU--- --(((((((((((....)))(((((..--....)))))...)))(((((((((.((....))))))))))))))))......((((.....))))...--- ( -36.70, z-score = -1.74, R) >droEre2.scaffold_4690 4976813 94 - 18748788 --UUGUGGGAGGGGCAACUCGCAUUUC--CUAUAAUGCAUUUCCGAGCUGCAGCUGCAGCAACUGCAGUUAUACAGUGAAAACCCCUCUACGGGGAUU--- --...((((((((....)))(((((..--....)))))..))))).((((.((((((((...))))))))...)))).....((((.....))))...--- ( -33.90, z-score = -2.40, R) >droWil1.scaffold_180777 457288 97 - 4753960 --UUGUGGGAGUGGCAAUUGCAAUUGCAACUAGGAUAAUUAAUUAAG--ACGAAUGCACCGCCAAUCCCCGAACACUUUUCAUUAUUCUCCACAAAUGACA --(((.((((.((((..((((....))))...((...(((.......--...)))...)))))).))))))).......(((((..........))))).. ( -19.30, z-score = -0.70, R) >consensus __GUGUGGGAGGGGCAACUCGCAUUUC__CCAGGAUGCAUUUCUGAGCUGCAGCUGCACCAACAGCAGUUCUACAGUGAAAACCCCUCAGCGGGGAUU___ ........((((((......(((((........)))))......(((((((.............)))))))...........))))))............. (-19.14 = -18.84 + -0.30)
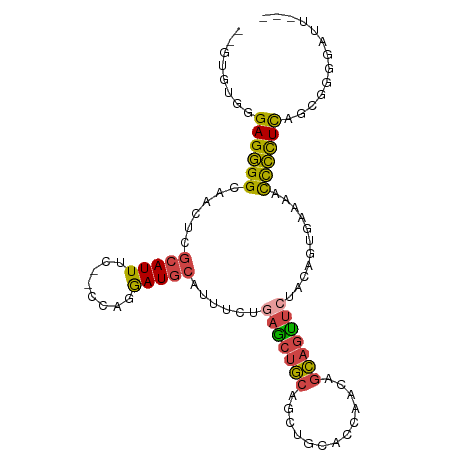
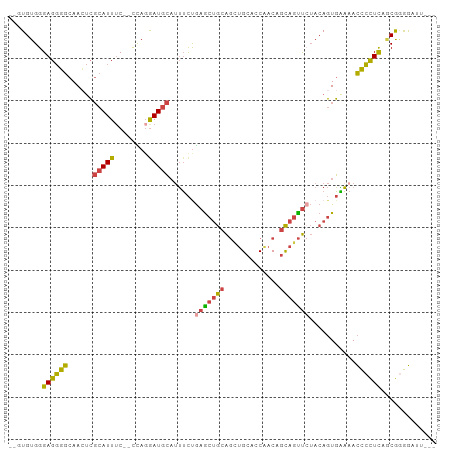
| Location | 13,103,938 – 13,104,034 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 73.13 |
| Shannon entropy | 0.49464 |
| G+C content | 0.49949 |
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -14.24 |
| Energy contribution | -13.83 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.762105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13103938 96 - 22422827 ---AAUCCCCGCUAAGGGGUUUCCACUGUAGAACUGCAGUUGGUGCAGCUGCAGCUCAGAAAUGCAUCCUGA--GAAAUGCGAGUUGCCCCUCCCACACAA ---...((((.....)))).....((((((....)))))).((.(((((((((.(((((.........))))--)...))).)))))).)).......... ( -33.70, z-score = -2.40, R) >droSim1.chrX 9993310 96 - 17042790 ---GAUCCCCGCUAAGGGGUUUUCACUGUAGAACUGCUGUUGGUGCAACUGCAGCUCAGAAAUGCAUCCUGA--GAAAUGCGUGUUGCCCCUCCCACACAA ---((.((((.....))))...))((.(((....))).)).((.(((((((((.(((((.........))))--)...)))).))))).)).......... ( -29.80, z-score = -1.57, R) >droSec1.super_21 849369 96 - 1102487 ---AAUCCUCGCUAAGGGGUUUUCACUGUAGAACUGCUGUUGGUGCAACUGCAGCUCAGAAAUGCAUCCUGA--GAAAUGCGAUUUGCCCCUCCCACACAA ---...........((((((..((.((((((...(((.......))).))))))(((((.........))))--)......))...))))))......... ( -25.90, z-score = -0.79, R) >droYak2.chrX 7375076 94 - 21770863 ---AAUCCCCGCAGAGGGGUUUUCACUGUAGAACUGCAGUGGCUGCAGCUGCAGUUCGGAACUGCAUCCUAG--GAAAUGCGAGUUGCCCCUCCCACAA-- ---..........(((((((.(((......((((((((((.......))))))))))......(((((....--))..))))))..)))))))......-- ( -35.30, z-score = -1.89, R) >droEre2.scaffold_4690 4976813 94 + 18748788 ---AAUCCCCGUAGAGGGGUUUUCACUGUAUAACUGCAGUUGCUGCAGCUGCAGCUCGGAAAUGCAUUAUAG--GAAAUGCGAGUUGCCCCUCCCACAA-- ---.......((.(((((((.(((.(((.....(((((((((...)))))))))..)))....(((((....--..))))))))..)))))))..))..-- ( -34.00, z-score = -2.70, R) >droWil1.scaffold_180777 457288 97 + 4753960 UGUCAUUUGUGGAGAAUAAUGAAAAGUGUUCGGGGAUUGGCGGUGCAUUCGU--CUUAAUUAAUUAUCCUAGUUGCAAUUGCAAUUGCCACUCCCACAA-- ......((((((.(((((.(....).)))))((((((.(((((.....))))--)..........))))(((((((....)))))))))....))))))-- ( -24.70, z-score = -0.96, R) >consensus ___AAUCCCCGCAAAGGGGUUUUCACUGUAGAACUGCAGUUGGUGCAGCUGCAGCUCAGAAAUGCAUCCUAA__GAAAUGCGAGUUGCCCCUCCCACAA__ ..............((((((.....(((.....((((.((((...)))).))))..)))....((((..........)))).....))))))......... (-14.24 = -13.83 + -0.41)

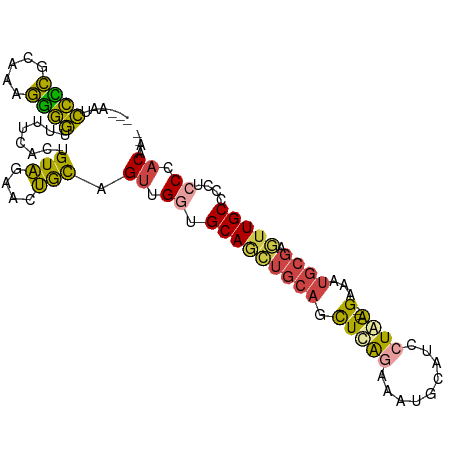
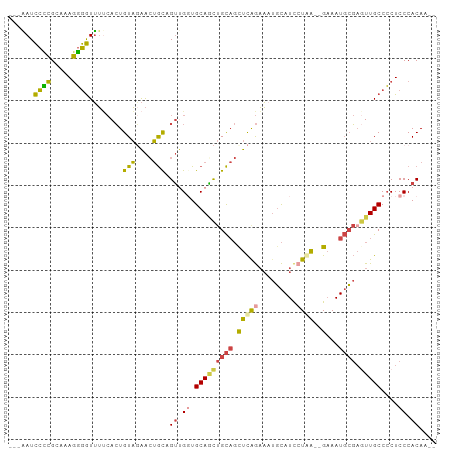
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:39:30 2011