
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,102,432 – 13,102,514 |
| Length | 82 |
| Max. P | 0.742985 |

| Location | 13,102,432 – 13,102,514 |
|---|---|
| Length | 82 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 65.50 |
| Shannon entropy | 0.64729 |
| G+C content | 0.54845 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -12.77 |
| Energy contribution | -12.83 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
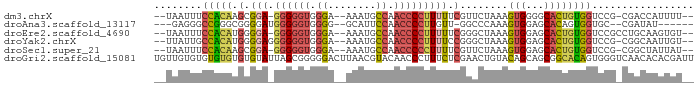
>dm3.chrX 13102432 82 - 22422827 --UAAUUUCCACAAGCGGA-GGGGGUGGGA--AAAUGCCAACCCCUUUUUCGUUCUAAAGUGGGGCACUGUGGUCCG-CGACCAUUUU-- --....((((((.((((((-((((((.((.--.....)).))))))..)))))).....))))))....((((((..-.))))))...-- ( -27.40, z-score = -0.81, R) >droAna3.scaffold_13117 2417238 76 - 5790199 ---GAGGGCCGGGCGGGGAUGGGGGUGGGG--GCAUUCCAACCCCUUGUU-GGCCCAAAGUGGAGCACAGUGGUGC--CGAUAU------ ---..(((((((..((((...((((((...--.))))))..))))...))-)))))...((.(.((((....))))--).))..------ ( -33.30, z-score = -0.75, R) >droEre2.scaffold_4690 4975314 83 + 18748788 --UAAUUUCCACAUGGGGA-GGGGGUGGGA--AAAUGCCAACCCCUUUUUCGGGCUAAAGUGGAGCACUGUGGUCCGCCUGCAAGUGU-- --.......(((..(..((-((((((.((.--.....)).))))))))..)((((((.((((...)))).))))))........))).-- ( -33.30, z-score = -1.67, R) >droYak2.chrX 7373496 83 - 21770863 --UUAUUGCCACAUGGGGAGGGGGGUGGGA--AAAUGCCAACCCCUUUUCCGGGCUAAAGUGGAGCACUGUGGUCCG-CGGCAAUUGU-- --..((((((.(...(((((((((((.((.--.....)).)))))))))))((((((.((((...)))).)))))))-.))))))...-- ( -41.00, z-score = -3.62, R) >droSec1.super_21 847893 82 - 1102487 --UAAUUUCCACAAGCGGA-GGGGGUGGGA--AAAUGCCAACCCCCUUUUCGUUCUAAAGUGGAGCACUGUGGUCCG-CGGCUAUUAU-- --((((..((...((((((-((((((.((.--.....)).))))))))..)))).....(((((.(.....).))))-)))..)))).-- ( -30.70, z-score = -2.06, R) >droGri2.scaffold_15081 2766737 90 - 4274704 UGUUGUGUGUGUGUGUGUAUUAGCGGGGGACUUAACGUACAACCCUUUCUCGAACUGUACAGCAGCGGCACAGUGGGUCAACACACGAUU .(((((((((...((((..(((..((....)))))..))))((((...((.(..((((......))))..))).))))..))))))))). ( -24.30, z-score = 0.86, R) >consensus __UAAUUUCCACAUGGGGA_GGGGGUGGGA__AAAUGCCAACCCCUUUUUCGGGCUAAAGUGGAGCACUGUGGUCCG_CGACAAUUGU__ ........(((((.......((((((.((........)).)))))).............(((...))))))))................. (-12.77 = -12.83 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:39:28 2011