| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,292,802 – 11,292,938 |
| Length | 136 |
| Max. P | 0.831678 |

| Location | 11,292,802 – 11,292,938 |
|---|---|
| Length | 136 |
| Sequences | 9 |
| Columns | 147 |
| Reading direction | forward |
| Mean pairwise identity | 74.63 |
| Shannon entropy | 0.52665 |
| G+C content | 0.43508 |
| Mean single sequence MFE | -34.09 |
| Consensus MFE | -20.96 |
| Energy contribution | -21.56 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.831678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
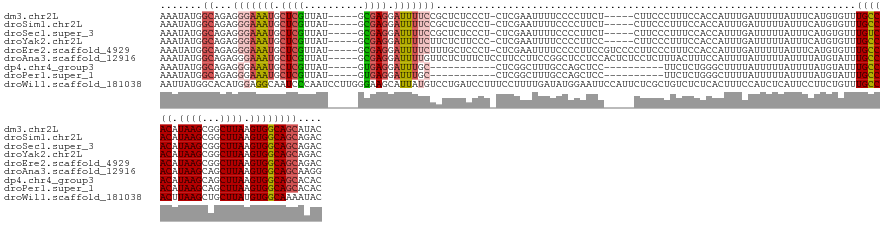
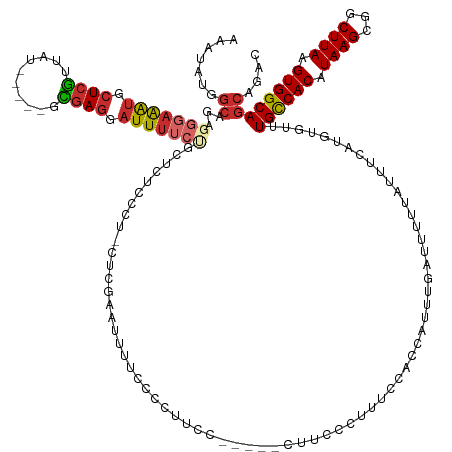
>dm3.chr2L 11292802 136 + 23011544 AAAUAUGGCAGAGGGAAAUGCUCGUUAU-----GCGAGGAUUUUCCGCUCUCCCU-CUCGAAUUUUCCCCUUCU-----CUUCCCUUUCCACCAUUUGAUUUUUAUUUCAUGUGUUUGCCACAUAAGCGGCUUAAGUGGCAGCAUAC .....(((.((((((((..((.......-----))((((.............)))-).................-----.))))))))))).....(((........)))((((((.(((((.((((...)))).))))))))))). ( -34.22, z-score = -1.54, R) >droSim1.chr2L 11096363 136 + 22036055 AAAUAUGGCAGAGGGAAAUGCUCGUUAU-----GCGAGGAUUUUCCGCUCUCCCU-CUCGAAUUUUCCCCUUCU-----CUUCCCUUUCCACCAUUUGAUUUUUAUUUCAUGUGUUUGCCACAUAAGCGGCUUAAGUGGCAGCAGAC .....(((.((((((((..((.......-----))((((.............)))-).................-----.))))))))))).(((.(((........))).))).(((((((.((((...)))).)))))))..... ( -33.32, z-score = -0.78, R) >droSec1.super_3 6689672 136 + 7220098 AAAUAUGGCAGAGGGAAAUGCUCGUUAU-----GCGAGGAUUUUCCGCUCUCCCU-CUCGAAUUUUCCCCUUCU-----CUUCCCUUUCCACCAUUUGAUUUUUAUUUCAUGUGUUUGUCACAUAAGCGGCUUAAGUGGCAGCAGAC .....(((.((((((((..((.......-----))((((.............)))-).................-----.))))))))))).(((.(((........))).))).(((((((.((((...)))).)))))))..... ( -30.62, z-score = -0.13, R) >droYak2.chr2L 7689321 136 + 22324452 AAAUAUGGCAGAGGGAAAUGCUCGUUAU-----GCGAGGAUUUUCUUCUCUUCCC-CUCGAAUUUUCCCCUUCC-----CUUCCCUUUCCACCAUUUGAUUUUUAUUUCAUGUGUUUGCCACAUAAGCGGCUUAAGUGGCAGCAGAC .....(((.((((((((..((.......-----))((((...............)-)))...............-----.))))))))))).(((.(((........))).))).(((((((.((((...)))).)))))))..... ( -33.66, z-score = -1.30, R) >droEre2.scaffold_4929 12491336 141 - 26641161 AAAUAUGGCAGAGGGAAAUGCUCGUUAU-----GCGAGGAUUUUCUUUGCUCCCU-CUCGAAUUUUCCCCUUCCGUCCCCUUCCCUUUCCACCAUUUGAUUUUUAUUUCAUGUGUUUGCCACAUAAGCGGCUUAAGUGGCAGCAGAC ......((.((.((((((...(((....-----((((((.....)))))).....-..)))..)))))))).))(((...............(((.(((........))).))).(((((((.((((...)))).)))))))..))) ( -36.10, z-score = -1.42, R) >droAna3.scaffold_12916 12014030 142 - 16180835 AAAUAUGGCAGAGGGAAAUGCUCGUUAU-----GCGAGGAUUUUGUUCUCUUUCUCCUUCCUUCCGGCUCCUCCACUCUCCUCUUUACUUUCCAUUUUAUUUUUAUUUUAUGUAUUUGCCACAUAAGCAGCUUAAGUGGCAGCAAGG ......((.(((((((...((.((..(.-----(.(((((....(....)....))))).))..))))...))).)))))).............................(((...((((((.((((...)))).)))))))))... ( -29.70, z-score = -0.21, R) >dp4.chr4_group3 6717825 121 + 11692001 AAAUAUGGCAGAGGGAAAUGCUCGUUAU-----GUGAGGAUUUGC-----------CUCGGCUUUGCCAGCUCC----------UUCUCUGGGCUUUUAUUUUUAUUUUAUGUAUUUGCCACAUAAGCAGCUUAAGUGGCAGCACAC .....(((((((((.((((.((((....-----.)))).)))).)-----------))).....)))))((.((----------......))))................(((..(((((((.((((...)))).))))))).))). ( -36.70, z-score = -1.37, R) >droPer1.super_1 8166340 121 + 10282868 AAAUAUGGCAGAGGGAAAUGCUCGUUAU-----GUGAGGAUUUGC-----------CUCGGCUUUGCCAGCUCC----------UUCUCUGGGCUUUUAUUUUUAUUUUAUGUAUUUGCCACAUAAGCAGCUUAAGUGGCAGCACAC .....(((((((((.((((.((((....-----.)))).)))).)-----------))).....)))))((.((----------......))))................(((..(((((((.((((...)))).))))))).))). ( -36.70, z-score = -1.37, R) >droWil1.scaffold_181038 202433 147 - 637489 AAUUAUGGCACAUGGAGGCAAUCCCAAUCCUUGGGAAGCAUUAUGUCCUGAUCCUUUCCUUUUGAUAUGGAAUUCCAUUCUCGCUGUCUCUCACUUUCCAUCUCAUUCCUUCUGUUUGCCACUUAAGCUGCUUAUGUGGCAAAAUAC ....((((....(((((((..(((((.....)))))(((..........((.....)).....((.((((....))))..)))))))))).))....)))).............((((((((.((((...)))).)))))))).... ( -35.80, z-score = -1.86, R) >consensus AAAUAUGGCAGAGGGAAAUGCUCGUUAU_____GCGAGGAUUUUCUGCUCUCCCU_CUCGAAUUUUCCCCUUCC_____CUUCCCUUUCCACCAUUUGAUUUUUAUUUCAUGUGUUUGCCACAUAAGCGGCUUAAGUGGCAGCAGAC .......((...(((((((.(((((........))))).)))))))......................................................................((((((.((((...)))).)))))))).... (-20.96 = -21.56 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:32:00 2011