
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,032,080 – 13,032,154 |
| Length | 74 |
| Max. P | 0.581565 |

| Location | 13,032,080 – 13,032,154 |
|---|---|
| Length | 74 |
| Sequences | 8 |
| Columns | 77 |
| Reading direction | forward |
| Mean pairwise identity | 57.46 |
| Shannon entropy | 0.82178 |
| G+C content | 0.44470 |
| Mean single sequence MFE | -22.93 |
| Consensus MFE | -0.28 |
| Energy contribution | -0.31 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.70 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.01 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
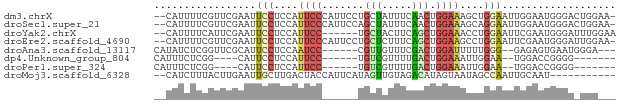
>dm3.chrX 13032080 74 + 22422827 --CAUUUUCGUUCGAAUUCCUCCAUUCCCAUUCCUGCUAUUUCAACUGGAAAGCUGGAAUUGGAAUGGGACUGGAA- --..................((((.(((((((((....((((((.((....)).)))))).))))))))).)))).- ( -27.90, z-score = -4.20, R) >droSec1.super_21 783982 74 + 1102487 --CAUUUUCGUUCGAAUUCCUCCAUUCCCAUUCCAGCUAUUUCAACUGGAAAGCAGGAAUUGGAAUGGGACUGGAA- --..................((((.((((((((((...(((((..((....))..))))))))))))))).)))).- ( -26.30, z-score = -3.31, R) >droYak2.chrX 7307545 69 + 21770863 --CAUUUUCAUUCGAAUUCCUCCAUUCC------UGCUACUUCAGCUGGAAACCUGGAAUUCGAAUGGGAUUUGGAA --((.(..((((((((((((....((((------.(((.....))).))))....))))))))))))..)..))... ( -28.20, z-score = -5.51, R) >droEre2.scaffold_4690 4907758 74 - 18748788 --CAUUUUCGUUCGAAUUCCUCCAUUCCCAUUCCUGCUCUUUCAGCUGGAAGCCUGGAAUUCGAAUGGGAUUGGAA- --((.(..((((((((((((..........((((.(((.....))).))))....))))))))))))..).))...- ( -26.64, z-score = -3.30, R) >droAna3.scaffold_13117 2357877 66 + 5790199 CAUAUCUCGGUUCGCAUUCCUCCAAUCC------CGUUGUUUCGACUGGAUUUUUGGG--GAGAGUGAAUGGGA--- ....(((((.(((((.((((((.(((((------.((((...)))).)))))...)))--))).))))))))))--- ( -23.90, z-score = -2.14, R) >dp4.Unknown_group_804 19328 58 - 58332 CAUUUCUCGG----CAUUCCUCCAUUCC------UGUCGUUUUGACUGGAAAUUGGAA--UGGACCGGGG------- .....(((((----((((((....((((------.(((.....))).))))...))))--))..))))).------- ( -21.30, z-score = -2.70, R) >droPer1.super_324 17941 58 + 26204 CAUUUCUCGG----CAUUCCUCCAUUCC------UGUCGUUUUGACUGGAAAUUGGAA--UGGACCGGGG------- .....(((((----((((((....((((------.(((.....))).))))...))))--))..))))).------- ( -21.30, z-score = -2.70, R) >droMoj3.scaffold_6328 2772333 64 - 4453435 --CAUCUUUACUUGAAUUGCUUGACUACCAUUCAUAGUUGUAGACAUAGUAAUAGCCAAUUGCAAU----------- --.............((((((((.((((.((.....)).)))).)).)))))).((.....))...----------- ( -7.90, z-score = 0.09, R) >consensus __CAUCUUCGUUCGAAUUCCUCCAUUCC______UGCUAUUUCAACUGGAAAUCUGGAAUUGGAAUGGGA_UGG___ .................(((....((((.......(((.....))).))))....)))................... ( -0.28 = -0.31 + 0.03)


| Location | 13,032,080 – 13,032,154 |
|---|---|
| Length | 74 |
| Sequences | 8 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 57.46 |
| Shannon entropy | 0.82178 |
| G+C content | 0.44470 |
| Mean single sequence MFE | -19.36 |
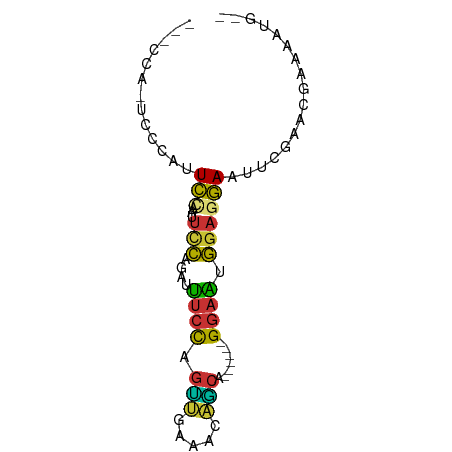
| Consensus MFE | -2.68 |
| Energy contribution | -1.76 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.90 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.14 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.581565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
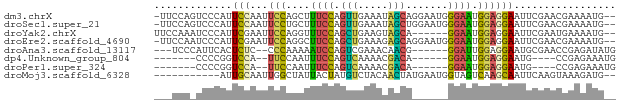
>dm3.chrX 13032080 74 - 22422827 -UUCCAGUCCCAUUCCAAUUCCAGCUUUCCAGUUGAAAUAGCAGGAAUGGGAAUGGAGGAAUUCGAACGAAAAUG-- -(((((.(((((((((.(((.(((((....))))).)))....))))))))).)))))...(((....)))....-- ( -24.80, z-score = -3.40, R) >droSec1.super_21 783982 74 - 1102487 -UUCCAGUCCCAUUCCAAUUCCUGCUUUCCAGUUGAAAUAGCUGGAAUGGGAAUGGAGGAAUUCGAACGAAAAUG-- -(((((.((((((((((....(((.((((.....))))))).)))))))))).)))))...(((....)))....-- ( -24.30, z-score = -2.89, R) >droYak2.chrX 7307545 69 - 21770863 UUCCAAAUCCCAUUCGAAUUCCAGGUUUCCAGCUGAAGUAGCA------GGAAUGGAGGAAUUCGAAUGAAAAUG-- ..........((((((((((((....((((.((((...)))).------))))....))))))))))))......-- ( -24.80, z-score = -4.27, R) >droEre2.scaffold_4690 4907758 74 + 18748788 -UUCCAAUCCCAUUCGAAUUCCAGGCUUCCAGCUGAAAGAGCAGGAAUGGGAAUGGAGGAAUUCGAACGAAAAUG-- -(((((.((((((((...(((..(((.....)))....)))...)))))))).)))))...(((....)))....-- ( -22.10, z-score = -1.80, R) >droAna3.scaffold_13117 2357877 66 - 5790199 ---UCCCAUUCACUCUC--CCCAAAAAUCCAGUCGAAACAACG------GGAUUGGAGGAAUGCGAACCGAGAUAUG ---((.(((((.((((.--......(((((.((.......)).------)))))))))))))).))........... ( -13.01, z-score = -0.15, R) >dp4.Unknown_group_804 19328 58 + 58332 -------CCCCGGUCCA--UUCCAAUUUCCAGUCAAAACGACA------GGAAUGGAGGAAUG----CCGAGAAAUG -------...(((..((--((((...((((.(((.....))).------))))....))))))----)))....... ( -17.40, z-score = -2.28, R) >droPer1.super_324 17941 58 - 26204 -------CCCCGGUCCA--UUCCAAUUUCCAGUCAAAACGACA------GGAAUGGAGGAAUG----CCGAGAAAUG -------...(((..((--((((...((((.(((.....))).------))))....))))))----)))....... ( -17.40, z-score = -2.28, R) >droMoj3.scaffold_6328 2772333 64 + 4453435 -----------AUUGCAAUUGGCUAUUACUAUGUCUACAACUAUGAAUGGUAGUCAAGCAAUUCAAGUAAAGAUG-- -----------(((((..((((((((((.((((........))))..))))))))))))))).............-- ( -11.10, z-score = -0.57, R) >consensus ___CCA_UCCCAUUCCAAUUCCAGAUUUCCAGUUGAAACAGCA______GGAAUGGAGGAAUUCGAACGAAAAUG__ ...................(((....((((.(((.....))).......))))....)))................. ( -2.68 = -1.76 + -0.92)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:39:19 2011