
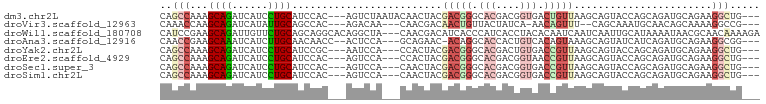
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,278,947 – 11,279,145 |
| Length | 198 |
| Max. P | 0.786044 |

| Location | 11,278,947 – 11,279,041 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 73.18 |
| Shannon entropy | 0.53948 |
| G+C content | 0.51529 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -9.96 |
| Energy contribution | -11.46 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.722039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11278947 94 + 23011544 CAGCCAAAGCAGAUCAUCCUGCAUCCAC---AGUCUAAUACAACUACGACGGGCACGACGGUGACUGUUAAGCAGUACCAGCAGAUGCAGAAGGCUG--- (((((......(.....)(((((((...---.((((..............)))).....((((.(((.....)))))))....)))))))..)))))--- ( -26.14, z-score = -1.39, R) >droVir3.scaffold_12963 17968979 88 - 20206255 CAAACCAAGCAGAUCAUAUUGCAGCCAC---AGACAA---CAACGACAACUGUUACUAUCA-AACAGUUU--CAGCAAAUGCAACAGCAAAAGGCCG--- ........((((......)))).(((..---......---....((.(((((((.......-))))))))--)......(((....)))...)))..--- ( -15.90, z-score = -2.25, R) >droWil1.scaffold_180708 8080062 97 - 12563649 CAUCCGAAGCAGAUUGUUCUGCAGCAGGCACAGGCUA---CAACGACAUCACCCAUCACCUACACAAUCAAUCAAUUGCAUAAAAUAACGCAACAAAAGA ........(((((....))))).((.(((....))).---........................((((......))))...........))......... ( -11.70, z-score = 0.12, R) >droAna3.scaffold_12916 12001436 91 - 16180835 CAACCGAAGCAAAUCAUCUUGCAACAACC--ACUCCA---GCAGAAC-ACAGGCACCACUGUCACAGUAAAGCAGUAUCAUCAGAUGCAGAAGGCGG--- ...(((..((((......)))).....((--......---((...((-((((......))))....))...)).(((((....)))))....)))))--- ( -16.70, z-score = -0.64, R) >droYak2.chr2L 7675498 91 + 22324452 CAGCCAAAGCAGAUCAUCCUGCAUCCGC---AAUCCA---CCACUACGACGGGCACGACUGUGACCGUUAAGCAGUACCAGCAGAUGCAGAAGGCUG--- (((((...((((......)))).((.((---(.((..---.......(((((.(((....))).)))))..((.......)).))))).)).)))))--- ( -28.00, z-score = -2.24, R) >droEre2.scaffold_4929 12474962 91 - 26641161 CAGCCAAAGCAGAUCAUCCUGCAUCCAC---AGUCCA---CCACUACGACGGGCACGACGGUAACCGUUAAGCAGUACCAGCAGAUGCAGAAGGCUG--- (((((......(.....)(((((((...---......---.......(((((..((....))..)))))..((.......)).)))))))..)))))--- ( -25.40, z-score = -1.86, R) >droSec1.super_3 6675764 91 + 7220098 CAGCCAAAGCAGAUCAUCCUGCAUCCAC---AGUCCA---CAACUACGACGGGCACGACGGUGACCGUUAAGCAGUACCAGCAGAUGCAGAAGGCUG--- (((((......(.....)(((((((...---......---.......(((((.(((....))).)))))..((.......)).)))))))..)))))--- ( -27.20, z-score = -2.02, R) >droSim1.chr2L 11082552 91 + 22036055 CAGCCAAAGCAGAUCAUCCUGCAUCCAC---AGUCCA---CAACUACGACGGGCACGACGGUGACCGUUAAGCAGUACCAGCAGAUGCAGAAGGCUG--- (((((......(.....)(((((((...---......---.......(((((.(((....))).)))))..((.......)).)))))))..)))))--- ( -27.20, z-score = -2.02, R) >consensus CAGCCAAAGCAGAUCAUCCUGCAUCCAC___AGUCCA___CAACUACGACGGGCACGACGGUGACCGUUAAGCAGUACCAGCAGAUGCAGAAGGCUG___ ..(((...((((......)))).........................(((((.(((....))).))))).......................)))..... ( -9.96 = -11.46 + 1.50)


| Location | 11,278,947 – 11,279,041 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 73.18 |
| Shannon entropy | 0.53948 |
| G+C content | 0.51529 |
| Mean single sequence MFE | -30.41 |
| Consensus MFE | -13.86 |
| Energy contribution | -14.81 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.63 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.786044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
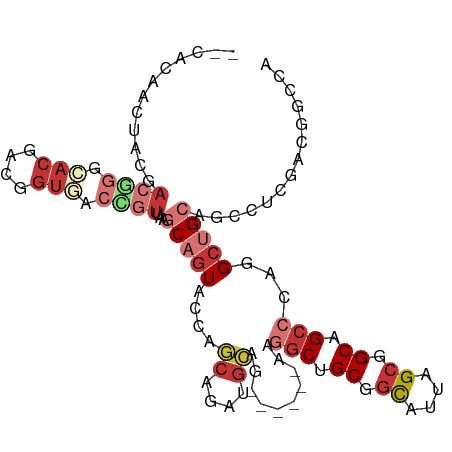
>dm3.chr2L 11278947 94 - 23011544 ---CAGCCUUCUGCAUCUGCUGGUACUGCUUAACAGUCACCGUCGUGCCCGUCGUAGUUGUAUUAGACU---GUGGAUGCAGGAUGAUCUGCUUUGGCUG ---((((((((((((((..(.(((((((.....)))).)))(((((((.(......)..))))..))).---)..)))))))))...........))))) ( -31.30, z-score = -1.43, R) >droVir3.scaffold_12963 17968979 88 + 20206255 ---CGGCCUUUUGCUGUUGCAUUUGCUG--AAACUGUU-UGAUAGUAACAGUUGUCGUUG---UUGUCU---GUGGCUGCAAUAUGAUCUGCUUGGUUUG ---.((((....((.(...(((((((.(--((((((((-.......))))))).))((..---(.....---)..)).)))).)))..).))..)))).. ( -18.60, z-score = 0.04, R) >droWil1.scaffold_180708 8080062 97 + 12563649 UCUUUUGUUGCGUUAUUUUAUGCAAUUGAUUGAUUGUGUAGGUGAUGGGUGAUGUCGUUG---UAGCCUGUGCCUGCUGCAGAACAAUCUGCUUCGGAUG ......(((((((......))))))).(((((.((((((((((.((((((.((......)---).)))))))))))).))))..)))))..(....)... ( -26.20, z-score = -1.38, R) >droAna3.scaffold_12916 12001436 91 + 16180835 ---CCGCCUUCUGCAUCUGAUGAUACUGCUUUACUGUGACAGUGGUGCCUGU-GUUCUGC---UGGAGU--GGUUGUUGCAAGAUGAUUUGCUUCGGUUG ---..(((....(((...(((.((..(((...((..(..(((..(.((....-)).)..)---))..).--.))....)))..)).))))))...))).. ( -22.20, z-score = 0.10, R) >droYak2.chr2L 7675498 91 - 22324452 ---CAGCCUUCUGCAUCUGCUGGUACUGCUUAACGGUCACAGUCGUGCCCGUCGUAGUGG---UGGAUU---GCGGAUGCAGGAUGAUCUGCUUUGGCUG ---(((((((((((((((((...(((..((..((((.(((....))).))))...))..)---))....---))))))))))))...........))))) ( -40.00, z-score = -3.27, R) >droEre2.scaffold_4929 12474962 91 + 26641161 ---CAGCCUUCUGCAUCUGCUGGUACUGCUUAACGGUUACCGUCGUGCCCGUCGUAGUGG---UGGACU---GUGGAUGCAGGAUGAUCUGCUUUGGCUG ---((((((((((((((..(.(.(((..((..((((.(((....))).))))...))..)---))..).---)..)))))))))...........))))) ( -35.80, z-score = -2.44, R) >droSec1.super_3 6675764 91 - 7220098 ---CAGCCUUCUGCAUCUGCUGGUACUGCUUAACGGUCACCGUCGUGCCCGUCGUAGUUG---UGGACU---GUGGAUGCAGGAUGAUCUGCUUUGGCUG ---((((((((((((((..(.(.(((.(((..((((.(((....))).))))...))).)---))..).---)..)))))))))...........))))) ( -34.60, z-score = -2.13, R) >droSim1.chr2L 11082552 91 - 22036055 ---CAGCCUUCUGCAUCUGCUGGUACUGCUUAACGGUCACCGUCGUGCCCGUCGUAGUUG---UGGACU---GUGGAUGCAGGAUGAUCUGCUUUGGCUG ---((((((((((((((..(.(.(((.(((..((((.(((....))).))))...))).)---))..).---)..)))))))))...........))))) ( -34.60, z-score = -2.13, R) >consensus ___CAGCCUUCUGCAUCUGCUGGUACUGCUUAACGGUCACCGUCGUGCCCGUCGUAGUUG___UGGACU___GUGGAUGCAGGAUGAUCUGCUUUGGCUG ...(((((((((((((((((.(((((.((............)).))))).(((............)))....))))))))))))...........))))) (-13.86 = -14.81 + 0.96)


| Location | 11,278,980 – 11,279,080 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 70.02 |
| Shannon entropy | 0.60001 |
| G+C content | 0.59107 |
| Mean single sequence MFE | -33.76 |
| Consensus MFE | -13.84 |
| Energy contribution | -16.09 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.695727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11278980 100 + 23011544 AAUACAACUACGACGGGCACGACGGUGACUGUUAAGCAGUACCAGCAGAUGCAG------AAGGCUGCGGCAUUAGCGGCAGCCCAGGCUGCAGCCUCGACGGCCA ..........((.(((((.....((((.(((.....)))))))......(((((------..((((((.((....)).))))))....)))))))).)).)).... ( -40.70, z-score = -2.34, R) >droVir3.scaffold_12963 17969009 91 - 20206255 ---------ACAACAACGACAACUGUUACUAUCAAACAGUUUCAGCAAAUGCAACAGCAAAAGGCCGCCGCCCUGGCUGCUGCACAGGCAGCUGCAGCUG------ ---------........((.(((((((.......))))))))).((....))..((((....(((....)))...((.(((((....))))).)).))))------ ( -27.90, z-score = -0.93, R) >droWil1.scaffold_180708 8080097 98 - 12563649 --UACAACGACAUCACCCAUCACCUACACAAUCAAUCAAUUGCAUAAAAUAACGCAACAAAAGACAGCAAUGGCUGCAGCCGCCGCCGCUGCAGCUGCUA------ --.....................................((((..........))))........((((...((((((((.......)))))))))))).------ ( -19.70, z-score = -1.38, R) >droAna3.scaffold_12916 12001469 93 - 16180835 --CAGCAGAAC-ACAGGCACCACUGUCACAGUAAAGCAGUAUCAUCAGAUGCAG------AAGGCGGCGGCUUUGGCGGCAGCACAAGCGGCAGCAGCAGCA---- --..((.....-((((......)))).........((.(((((....)))))..------...((.((.((((..((....))..)))).)).)).)).)).---- ( -27.20, z-score = -0.63, R) >droYak2.chr2L 7675530 98 + 22324452 --CACCACUACGACGGGCACGACUGUGACCGUUAAGCAGUACCAGCAGAUGCAG------AAGGCUGCGGCAUUAGCGGCAGCCCAGGCUGCAGCCUCGACGGCCA --.........(((((.(((....))).)))))..(((((....((....))..------..((((((.((....)).))))))...))))).(((.....))).. ( -38.10, z-score = -1.30, R) >droEre2.scaffold_4929 12474994 98 - 26641161 --CACCACUACGACGGGCACGACGGUAACCGUUAAGCAGUACCAGCAGAUGCAG------AAGGCUGCGGCAUUAGCGGCAGCCCAGGCUGCGUCCUCGACGGCCA --........(((.((((..(((((...)))))..(((((....((....))..------..((((((.((....)).))))))...))))))))))))....... ( -38.90, z-score = -1.73, R) >droSec1.super_3 6675796 98 + 7220098 --CACAACUACGACGGGCACGACGGUGACCGUUAAGCAGUACCAGCAGAUGCAG------AAGGCUGCGGCAUUAGCGGCAGCCCAGGCUGCAGCCUCGACGGCCA --........((.(((((..(((((...)))))..(((((....((....))..------..((((((.((....)).))))))...))))).))).)).)).... ( -38.80, z-score = -1.73, R) >droSim1.chr2L 11082584 98 + 22036055 --CACAACUACGACGGGCACGACGGUGACCGUUAAGCAGUACCAGCAGAUGCAG------AAGGCUGCGGCACUAGCGGCAGCCCAGGCUGCAGCCUCGACGGCCA --........((.(((((..(((((...)))))..(((((....((....))..------..((((((.((....)).))))))...))))).))).)).)).... ( -38.80, z-score = -1.41, R) >consensus __CACAACUACGACGGGCACGACGGUGACCGUUAAGCAGUACCAGCAGAUGCAG______AAGGCUGCGGCAUUAGCGGCAGCCCAGGCUGCAGCCUCGACGGCCA ............((((.(((....))).))))...(((((....((....))..........(((.((.((....)).)).)))...))))).............. (-13.84 = -16.09 + 2.25)


| Location | 11,279,041 – 11,279,145 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.05 |
| Shannon entropy | 0.47005 |
| G+C content | 0.59254 |
| Mean single sequence MFE | -41.39 |
| Consensus MFE | -19.38 |
| Energy contribution | -17.10 |
| Covariance contribution | -2.28 |
| Combinations/Pair | 1.68 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
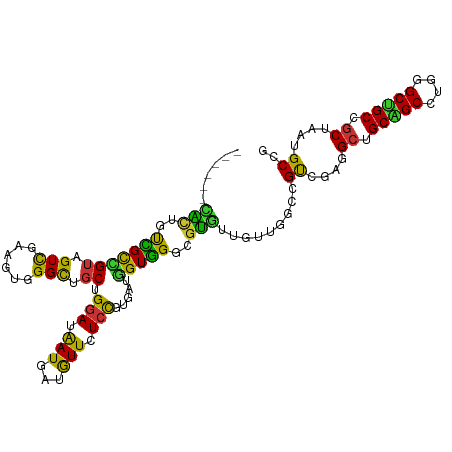
>dm3.chr2L 11279041 104 - 23011544 ------CACUGUCGCCGUAGUCGAAGUGGGCUGCUGGAUAAUGAUGUUCUCCGUGAUGGUGGGCGUGUUGUUGGCCGUCGAGGCUGCAGCCUGGGCUGCCGCUAAUGCCG ------(((((((((.((((((......)))))).(((.(((...))).))))))))))))(((((...((.(((.(((.((((....)))).))).)))))..))))). ( -47.00, z-score = -2.66, R) >droVir3.scaffold_12963 17969067 98 + 20206255 ------CGUCACGGCUGUCGUUGAUGUGGGUUGCUGUAUGAUGAUGUUUUCUGUGAUGGUUGGUGCAUUG------GCAGCUGCAGCUGCCUGUGCAGCAGCCAGGGCGG ------.(((((((.(((((((.(((..(....)..))))))))))....)))))))(((((.(((((.(------(((((....)))))).))))).)))))....... ( -43.40, z-score = -2.36, R) >droWil1.scaffold_180708 8080159 107 + 12563649 CGUUGGUGACGUGGUCGUGGUUUGUGGUGGCGGCUGGAUGAUGAUAUUUUCCGUAAUCGUUGGAGUAUUACUA---GCAGCUGCAGCGGCGGCGGCUGCAGCCAUUGCUG ...((((...(..(((((.(((.((.(..((.(((((.(((((...(((..((....))..))).))))))))---)).))..).))))).)))))..).))))...... ( -39.20, z-score = -0.86, R) >droAna3.scaffold_12916 12001527 98 + 16180835 ------UGCUGUUGUGGAAGUUGAAGUGGGCUGCUGGAUAAUGAUGUUUUCCGUGAUUGUGGGUGUGUU------UGCUGCUGCUGCCGCUUGUGCUGCCGCCAAAGCCG ------.(((...((((.(((..((((((((.((...(((((.(((.....))).))))).(((.....------.))))).))..))))))..))).))))...))).. ( -27.60, z-score = 0.06, R) >droYak2.chr2L 7675589 104 - 22324452 ------CACUGCCGCCGUUGUCGAAGUGGGUUGCUGGAUAAUGAUGUUCUCCGUAAUGGUGGGCGUGUUGUUGGCCGUCGAGGCUGCAGCCUGGGCUGCCGCUAAUGCCG ------((((.((((((....))..))))(((((.(((.(((...))).))))))))))))(((((...((.(((.(((.((((....)))).))).)))))..))))). ( -39.10, z-score = -0.46, R) >droEre2.scaffold_4929 12475053 104 + 26641161 ------CACUGUCGCUGUAGUCGAAGUGGGCUGCUGGAUAAUGAUGUUCUCCGUGAUGGUGGGCGUGUUGUUGGCCGUCGAGGACGCAGCCUGGGCUGCCGCUAAUGCCG ------(((((((((.((((((......)))))).(((.(((...))).))))))))))))(((((...((.(((.(((.(((......))).))).)))))..))))). ( -43.50, z-score = -1.94, R) >droSec1.super_3 6675855 104 - 7220098 ------CACUGUCGCCGUAGUCGAAGUGGGCUGCUGGAUAAUGAUGUUCUCCGUGAUGGUGGGCGUGUUGUUGGCCGUCGAGGCUGCAGCCUGGGCUGCCGCUAAUGCCG ------(((((((((.((((((......)))))).(((.(((...))).))))))))))))(((((...((.(((.(((.((((....)))).))).)))))..))))). ( -47.00, z-score = -2.66, R) >droSim1.chr2L 11082643 104 - 22036055 ------CACUGUCGCCGUAGUCGAAGUGGGCUGCUGGAUAAUGAUGUUCUCCGUGAUGGUGGGCGUGUUGUUGGCCGUCGAGGCUGCAGCCUGGGCUGCCGCUAGUGCCG ------(((((((((.((((((......)))))).(((.(((...))).))))))))))))(((((...((.(((.(((.((((....)))).))).)))))..))))). ( -44.30, z-score = -1.54, R) >consensus ______CACUGUCGCCGUAGUCGAAGUGGGCUGCUGGAUAAUGAUGUUCUCCGUGAUGGUGGGCGUGUUGUUGGCCGUCGAGGCUGCAGCCUGGGCUGCCGCUAAUGCCG ......(((..(((((((.(((......))).)).(((.(((...))).))).....)))))..))).........((....((.(((((....))))).))....)).. (-19.38 = -17.10 + -2.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:59 2011