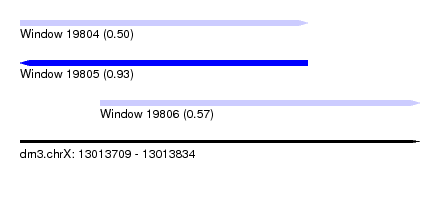
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,013,709 – 13,013,834 |
| Length | 125 |
| Max. P | 0.926790 |

| Location | 13,013,709 – 13,013,799 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 84.15 |
| Shannon entropy | 0.27206 |
| G+C content | 0.54800 |
| Mean single sequence MFE | -35.44 |
| Consensus MFE | -23.82 |
| Energy contribution | -25.14 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
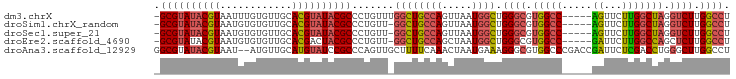
>dm3.chrX 13013709 90 + 22422827 -GCGUAUACGUAAUUUGUGUUGCACGUAUACGCCCUGUUUGGCUGCCAGUUAAUGGCUGGGCGUGGCC-----AGUUCUUGGCUAGGUCUUGGCCU -(((((((((((((....)))..)))))))))).......((((((((.....)))).((((.(((((-----((...))))))).)))).)))). ( -37.50, z-score = -2.48, R) >droSim1.chrX_random 3593493 89 + 5698898 -GCGUAUACGUAAUGUGUGUUGCACGUAUACGCCCUGUU-GGCUGCCAGUUAAUGGCUGGGCGUGGCC-----AGUUCUUGGCUAGGUCUUGGCCU -((((((((((...((.....))))))))))))......-((((((((.....)))).((((.(((((-----((...))))))).)))).)))). ( -37.70, z-score = -2.09, R) >droSec1.super_21 765526 89 + 1102487 -GCGUAUACGUAAUGUGUGUUGCACGUAUACGCCCUGUU-GGCUGCCAGUUAAUGGCUGGGCGUGGCC-----AGUUCUUGGCUAGGUCUUGGCCU -((((((((((...((.....))))))))))))......-((((((((.....)))).((((.(((((-----((...))))))).)))).)))). ( -37.70, z-score = -2.09, R) >droEre2.scaffold_4690 4889529 89 - 18748788 -GCGUAUACGUAAUGUGUGUUGCACGACUACGCCCUGUU-GGCUGCCAGCUAAUGGCUGGGCGUGGCC-----GAUUCUUGGCCAGCUCUUGGCCU -(((.(((((.....)))))))).((.((((((((.(((-(((.....))))))....)))))))).)-----)......((((((...)))))). ( -35.40, z-score = -1.26, R) >droAna3.scaffold_12929 2500482 94 + 3277472 GGCGUAUACGUAAU--AUGUUGCAUGUAUCCGCCCAGUUGCUUUUCAAACUAAUGAAAGGGCGUGGCCCGACCGAUUCUCGACCUGGGCUUGGCCU ((((((((.((((.--...)))).)))))..(((((((((..(((((......)))))((((...))))..........))).))))))...))). ( -28.90, z-score = -0.81, R) >consensus _GCGUAUACGUAAUGUGUGUUGCACGUAUACGCCCUGUU_GGCUGCCAGUUAAUGGCUGGGCGUGGCC_____AGUUCUUGGCUAGGUCUUGGCCU .((((((((((............)))))))))).......((((((((.....)))).((((.(((((............))))).)))).)))). (-23.82 = -25.14 + 1.32)
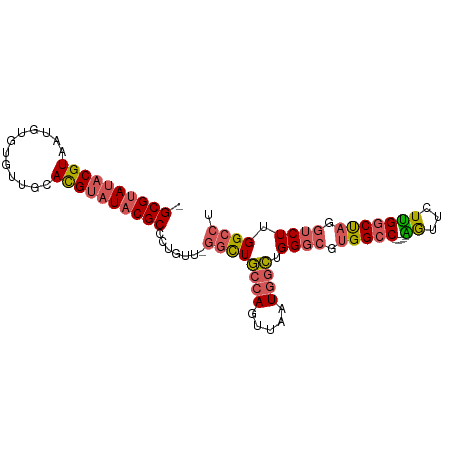
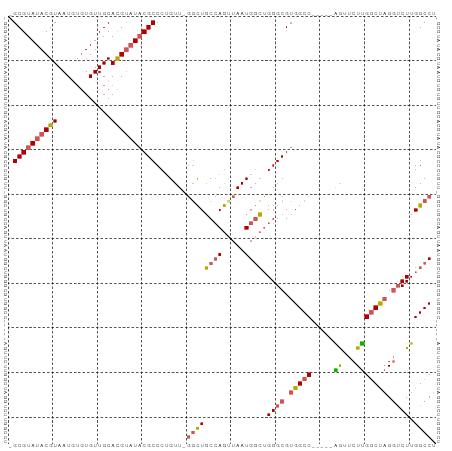
| Location | 13,013,709 – 13,013,799 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 84.15 |
| Shannon entropy | 0.27206 |
| G+C content | 0.54800 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -21.84 |
| Energy contribution | -21.84 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.926790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13013709 90 - 22422827 AGGCCAAGACCUAGCCAAGAACU-----GGCCACGCCCAGCCAUUAACUGGCAGCCAAACAGGGCGUAUACGUGCAACACAAAUUACGUAUACGC- .(((((.(..((.....))..))-----))))..((((.((((.....)))).(.....).))))(((((((((..........)))))))))..- ( -29.60, z-score = -2.75, R) >droSim1.chrX_random 3593493 89 - 5698898 AGGCCAAGACCUAGCCAAGAACU-----GGCCACGCCCAGCCAUUAACUGGCAGCC-AACAGGGCGUAUACGUGCAACACACAUUACGUAUACGC- .(((((.(..((.....))..))-----))))..((((.((((.....)))).(..-..).))))(((((((((..........)))))))))..- ( -29.60, z-score = -2.64, R) >droSec1.super_21 765526 89 - 1102487 AGGCCAAGACCUAGCCAAGAACU-----GGCCACGCCCAGCCAUUAACUGGCAGCC-AACAGGGCGUAUACGUGCAACACACAUUACGUAUACGC- .(((((.(..((.....))..))-----))))..((((.((((.....)))).(..-..).))))(((((((((..........)))))))))..- ( -29.60, z-score = -2.64, R) >droEre2.scaffold_4690 4889529 89 + 18748788 AGGCCAAGAGCUGGCCAAGAAUC-----GGCCACGCCCAGCCAUUAGCUGGCAGCC-AACAGGGCGUAGUCGUGCAACACACAUUACGUAUACGC- .(((.....(((((((.......-----))))).))(((((.....)))))..)))-....(.(((((((.(((.....))))))))))...)..- ( -31.60, z-score = -1.99, R) >droAna3.scaffold_12929 2500482 94 - 3277472 AGGCCAAGCCCAGGUCGAGAAUCGGUCGGGCCACGCCCUUUCAUUAGUUUGAAAAGCAACUGGGCGGAUACAUGCAACAU--AUUACGUAUACGCC .(((...((((.(..((.....))..)))))..((((((((((......))))).......)))))..............--...........))) ( -27.11, z-score = -0.87, R) >consensus AGGCCAAGACCUAGCCAAGAACU_____GGCCACGCCCAGCCAUUAACUGGCAGCC_AACAGGGCGUAUACGUGCAACACACAUUACGUAUACGC_ .((((.......((.......)).....))))..((((.((((.....)))).(.....).))))(((((((((..........)))))))))... (-21.84 = -21.84 + -0.00)
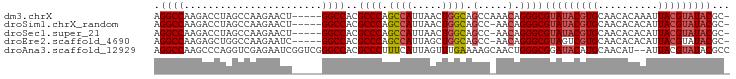
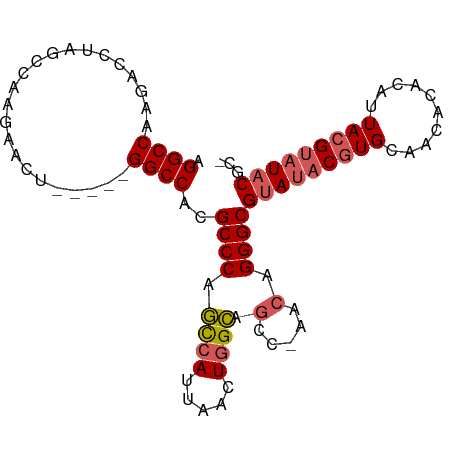
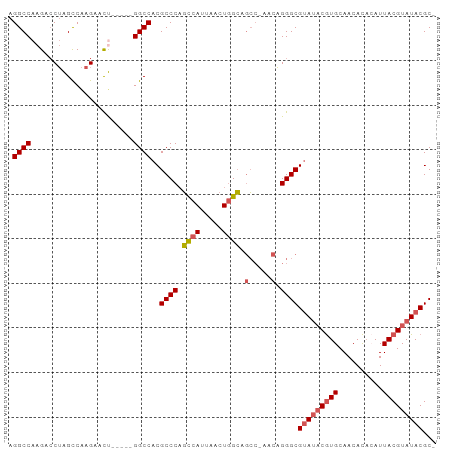
| Location | 13,013,734 – 13,013,834 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.91 |
| Shannon entropy | 0.40089 |
| G+C content | 0.52256 |
| Mean single sequence MFE | -35.57 |
| Consensus MFE | -18.11 |
| Energy contribution | -20.75 |
| Covariance contribution | 2.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.571883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
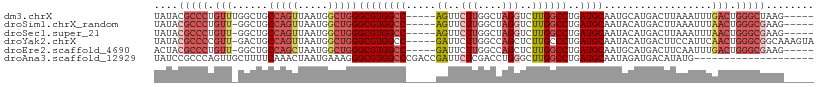
>dm3.chrX 13013734 100 + 22422827 UAUACGCCCUGUUUGGCUGCCAGUUAAUGGCUGGGCGUGGCC-----AGUUCUUGGCUAGGUCUUGGCCUGAUGCAAUGCAUGACUUAAAUUUGACUGGGCUAAG----- .....((((.(((.((((((((.....)))).((((.(((((-----((...))))))).)))).))))..((((...))))...........))).))))....----- ( -35.80, z-score = -1.32, R) >droSim1.chrX_random 3593518 99 + 5698898 UAUACGCCCUGUU-GGCUGCCAGUUAAUGGCUGGGCGUGGCC-----AGUUCUUGGCUAGGUCUUGGCCUGAUGCAAUACAUGACUUAAAUUUAACUGGGCGAAG----- ....(((((.(((-((((((((.....)))).((((.(((((-----((...))))))).)))).))))..(((.....)))...........))).)))))...----- ( -36.30, z-score = -1.91, R) >droSec1.super_21 765551 99 + 1102487 UAUACGCCCUGUU-GGCUGCCAGUUAAUGGCUGGGCGUGGCC-----AGUUCUUGGCUAGGUCUUGGCCUGAUGCAAUACAUGACUUAAAUUUAACUGGGCGAAG----- ....(((((.(((-((((((((.....)))).((((.(((((-----((...))))))).)))).))))..(((.....)))...........))).)))))...----- ( -36.30, z-score = -1.91, R) >droYak2.chrX 7289178 104 + 21770863 UAUACGCCCCGUU-GACUGCCAGUUAAUGGCUGGGCGUGGCC-----GAUUCUUGGCCAGCUCUUGCCCUGAUGCAAUACAUGACUUCCAUUCAACUGGGCGGCAAAGUA ....(((((.(((-((.((..((((...(((.((((.(((((-----((...)))))))))))..)))...(((.....)))))))..)).))))).)))))........ ( -40.80, z-score = -2.65, R) >droEre2.scaffold_4690 4889554 99 - 18748788 ACUACGCCCUGUU-GGCUGCCAGCUAAUGGCUGGGCGUGGCC-----GAUUCUUGGCCAGCUCUUGGCCUGAUGCAAUGCAUGACUUCAAUUUGACUGGGCGAAG----- ....(((((.(((-((((((((.....)))).((((.(((((-----((...)))))))))))..))))..((((...))))...........))).)))))...----- ( -40.80, z-score = -2.56, R) >droAna3.scaffold_12929 2500506 90 + 3277472 UAUCCGCCCAGUUGCUUUUCAAACUAAUGAAAGGGCGUGGCCCGACCGAUUCUCGACCUGGGCUUGGCCUGAUGCAAUAGAUGACAUAUG-------------------- ..........(((((.(((((......)))))((((..((((((..((.....))...))))))..))))...)))))............-------------------- ( -23.40, z-score = -0.45, R) >consensus UAUACGCCCUGUU_GGCUGCCAGUUAAUGGCUGGGCGUGGCC_____AAUUCUUGGCCAGGUCUUGGCCUGAUGCAAUACAUGACUUAAAUUUAACUGGGCGAAG_____ ....(((((.(((......(((((.....)))))((((((((............))))((((....)))).))))..................))).)))))........ (-18.11 = -20.75 + 2.64)
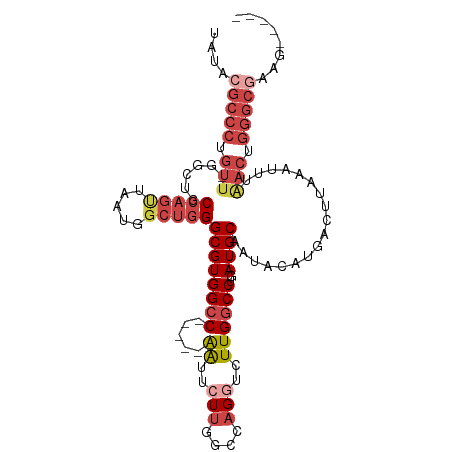
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:39:12 2011