
| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,012,601 – 13,012,777 |
| Length | 176 |
| Max. P | 0.860819 |

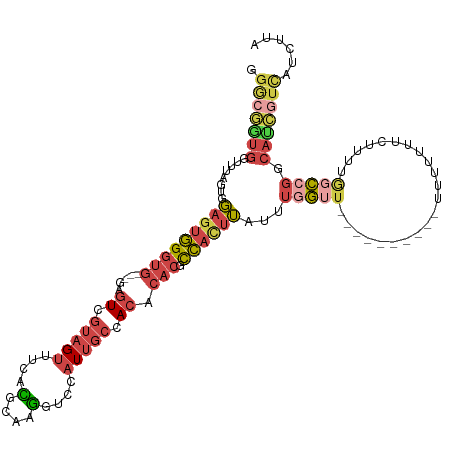
| Location | 13,012,601 – 13,012,718 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.94 |
| Shannon entropy | 0.62957 |
| G+C content | 0.46154 |
| Mean single sequence MFE | -27.46 |
| Consensus MFE | -13.28 |
| Energy contribution | -13.90 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.52 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.662669 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 13012601 117 + 22422827 GGACGGUGGUUAAGUGGAGUGGGUG---GAGUCGUAGUUUCACGCAAGGUCCAUUGCCACACACGCCAUUUAUUUUGUUGUUUUUCUUUUUUUUUUCUUUUGGCCGGCAUCGUCAUCUUA .(((((((((..((((((.((.(((---(((......)))))).))...)))))))))))....((((................................))))......))))...... ( -30.05, z-score = -1.17, R) >droSim1.chrU 148635 91 - 15797150 GGGCGGUGGUUUUGUGGAGUGGGUG---GAGUCGUAGUUUCACGCAAGGUCCAUUGCCACACACGCCACUUAUUUGG--------------------------CCGGCAUCGUCAUCUUA .(((((((((...(((((.((.(((---(((......)))))).))...))))).)))))....((((......)))--------------------------)......))))...... ( -31.40, z-score = -1.27, R) >droSec1.super_21 756340 111 + 1102487 GGGCGGUGGUUUUGUGGAGUGGGUG---GAGUCGUAGUUUCACGCAAGGUCCAUUGCCACACACGCCAGUUAUUUGGUUGUUU------UUUUGUUUCUUUGGCCGGCAUCGUCAUCUUA .(((((((((...(((((.((.(((---(((......)))))).))...))))).)))))....(((.((((...((......------.......))..)))).)))..))))...... ( -33.22, z-score = -1.32, R) >droYak2.chrX 7288155 93 + 21770863 --------------UGGAGUGGGUG---GAGUCGUAGUUUCAUGCAAGGUCCAUUGCCACACACGCCACUUAUUUGGUU----------UUUAUUUCUUGUGGCCGGCAUCGUCAUCUUA --------------((((.((.(((---(((......)))))).))...)))).((((......(((((..........----------..........))))).))))........... ( -20.05, z-score = 0.94, R) >droEre2.scaffold_4690 4888459 107 - 18748788 GGGCGAUGGUUAAGUGGAGUGGGUG---GAGUUGUAGUUUCACGCAAGGUCCAUUGCCACACACGCCACUUAUUUGGUU----------UUUAUUUUCUUUGGCCGGCAUCGUCGUCUUA (((((((((((((((((.(((.(((---(((......))))))((((......))))....))).))))))).((((((----------............)))))).)))))))))).. ( -35.40, z-score = -2.30, R) >triCas2.ChLG5 8074508 106 - 18847211 GGACAUUGUCUUAUUUCAGCAGAGAUCCGCCUUUUAGAUACAUGCAGAUUUUAUGUUAACACACCUUUCCGAUGAUAAUG----------CUUUUCAUUAUAAUUGACAAUAUUUA---- ....((((((.......((((((((((.((.............)).)))))).)))).............(((.((((((----------.....)))))).))))))))).....---- ( -14.62, z-score = -0.35, R) >consensus GGGCGGUGGUUUAGUGGAGUGGGUG___GAGUCGUAGUUUCACGCAAGGUCCAUUGCCACACACGCCACUUAUUUGGUU__________UUUUUUUCUUUUGGCCGGCAUCGUCAUCUUA .(((((((.....((((((((((.....(((.......))).(....).)))))).))))....((((......)))).............................)))))))...... (-13.28 = -13.90 + 0.62)

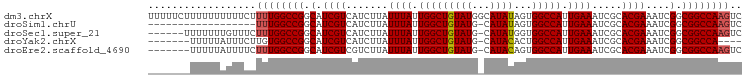
| Location | 13,012,678 – 13,012,777 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 86.14 |
| Shannon entropy | 0.24001 |
| G+C content | 0.44672 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -22.42 |
| Energy contribution | -22.58 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.860819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13012678 99 + 22422827 UUUUUCUUUUUUUUUUCUUUUGGCCGGCAUCGUCAUCUUAUUUAUUGGCUGUAUGGCAUAUAGUGGCCAUUGAAAUCGCACGAAAUCGGCGGCCAAGUC ..................((((((((.(.((((.......((((.((((..(..........)..)))).)))).....))))....).)))))))).. ( -24.20, z-score = -1.23, R) >droSim1.chrU 148704 80 - 15797150 ------------------UUUGGCCGGCAUCGUCAUCUUAUUUAUUGGCUGUAUG-CAUAUAGUGGCCAUUGAAAUCGCACGAAAUCGGCGGCCAAGUC ------------------((((((((.(.((((.......((((.((((..(...-......)..)))).)))).....))))....).)))))))).. ( -24.10, z-score = -1.35, R) >droSec1.super_21 756417 92 + 1102487 ------UUUUUUUGUUUCUUUGGCCGGCAUCGUCAUCUUAUUUAUUGGCUGUAUG-CAUAUGGUGGCCAUUGAAAUCGCACGAAAUCGGCGGCCAAGUC ------.......(((((..((((((.(((.((((..........))))((....-)).))).))))))..)))))(((.((....)))))........ ( -24.90, z-score = -0.97, R) >droYak2.chrX 7288215 87 + 21770863 -------UUUUUAUUUCUUGUGGCCGGCAUCGUCAUCUUAUUUAUUGGCUGUAUG-CAUACACUGGCCAUUGAAAUCGCACGAAAUCGGCGGCCA---- -------.....(((((..((((((((....((((..........))))((((..-..)))))))))))).)))))(((.((....)))))....---- ( -24.60, z-score = -1.55, R) >droEre2.scaffold_4690 4888533 91 - 18748788 -------UUUUUAUUUUCUUUGGCCGGCAUCGUCGUCUUAUUUAUUGGCUGUAUG-CAUACAGUGGCCAUUGAAAUCGCACGAAAUCGGCGGCCAAGUC -------...........((((((((.(....(((((...((((.((((..(.((-....)))..)))).))))...).))))....).)))))))).. ( -27.60, z-score = -2.27, R) >consensus _______UUUUUAUUUCCUUUGGCCGGCAUCGUCAUCUUAUUUAUUGGCUGUAUG_CAUAUAGUGGCCAUUGAAAUCGCACGAAAUCGGCGGCCAAGUC ..................((((((((.(.((((.......((((.(((((((((...))))...))))).)))).....))))....).)))))))).. (-22.42 = -22.58 + 0.16)


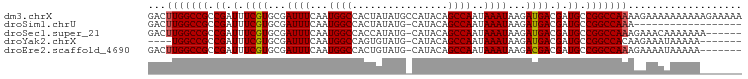
| Location | 13,012,678 – 13,012,777 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 86.14 |
| Shannon entropy | 0.24001 |
| G+C content | 0.44672 |
| Mean single sequence MFE | -20.24 |
| Consensus MFE | -18.25 |
| Energy contribution | -18.29 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.576039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 13012678 99 - 22422827 GACUUGGCCGCCGAUUUCGUGCGAUUUCAAUGGCCACUAUAUGCCAUACAGCCAAUAAAUAAGAUGACGAUGCCGGCCAAAAGAAAAAAAAAAGAAAAA ...(((((((.(((..((....))..)).(((((........)))))........................).)))))))................... ( -20.70, z-score = -1.44, R) >droSim1.chrU 148704 80 + 15797150 GACUUGGCCGCCGAUUUCGUGCGAUUUCAAUGGCCACUAUAUG-CAUACAGCCAAUAAAUAAGAUGACGAUGCCGGCCAAA------------------ ...(((((((.(((..((....))..))..((((...(((...-.)))..)))).................).))))))).------------------ ( -18.60, z-score = -0.50, R) >droSec1.super_21 756417 92 - 1102487 GACUUGGCCGCCGAUUUCGUGCGAUUUCAAUGGCCACCAUAUG-CAUACAGCCAAUAAAUAAGAUGACGAUGCCGGCCAAAGAAACAAAAAAA------ ...(((..(((((....)).))).((((..(((((..(((...-(((................)))...)))..)))))..))))))).....------ ( -18.79, z-score = -0.61, R) >droYak2.chrX 7288215 87 - 21770863 ----UGGCCGCCGAUUUCGUGCGAUUUCAAUGGCCAGUGUAUG-CAUACAGCCAAUAAAUAAGAUGACGAUGCCGGCCACAAGAAAUAAAAA------- ----((((((.(((..((....))..))..((((..((((...-.)))).)))).................).)))))).............------- ( -20.40, z-score = -0.65, R) >droEre2.scaffold_4690 4888533 91 + 18748788 GACUUGGCCGCCGAUUUCGUGCGAUUUCAAUGGCCACUGUAUG-CAUACAGCCAAUAAAUAAGACGACGAUGCCGGCCAAAGAAAAUAAAAA------- ...(((((((.((.(.((((...((((...((((...((((..-..))))))))..))))...)))).).)).)))))))............------- ( -22.70, z-score = -1.70, R) >consensus GACUUGGCCGCCGAUUUCGUGCGAUUUCAAUGGCCACUAUAUG_CAUACAGCCAAUAAAUAAGAUGACGAUGCCGGCCAAAAAAAAAAAAAA_______ ...(((((((.((.(.((((...((((...((((................))))..))))...)))).).)).)))))))................... (-18.25 = -18.29 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:39:09 2011