| Sequence ID | dm3.chrX |
|---|---|
| Location | 13,008,862 – 13,008,919 |
| Length | 57 |
| Max. P | 0.830319 |

| Location | 13,008,862 – 13,008,919 |
|---|---|
| Length | 57 |
| Sequences | 7 |
| Columns | 74 |
| Reading direction | forward |
| Mean pairwise identity | 55.52 |
| Shannon entropy | 0.84420 |
| G+C content | 0.40680 |
| Mean single sequence MFE | -11.76 |
| Consensus MFE | -5.11 |
| Energy contribution | -5.50 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.67 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
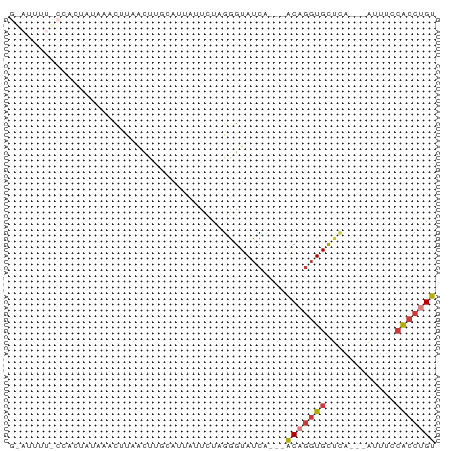
>dm3.chrX 13008862 57 + 22422827 GAAUUUUUCCAUUAUGAAUAU-----------UUCUAGGGUAUCA---ACAGGUGCUCA---AUUUCCACCUGU (((...(((......)))...-----------)))..........---(((((((....---.....))))))) ( -9.30, z-score = -0.56, R) >droPer1.super_21 268368 71 + 1645244 GGAUUUUCCUCGAAAAACCGUCACUAGAAAGCGUAUCGUGUAACACUCACACGUGCCUC---GUUUUGGGAAGU ((.(((((...))))).)).((.((((((((.(((.(((((.......)))))))))).---.))))))))... ( -14.60, z-score = -0.04, R) >droAna3.scaffold_12929 2496686 62 + 3277472 ---------GACUUUAGUCAUUGCUGUCAUUAGUGUAGGGUAUCA---ACAGGUGUCUACCUGUUCUCGCCUGU ---------(((....)))...((((....))))...((((...(---((((((....)))))))...)))).. ( -16.30, z-score = -1.49, R) >droEre2.scaffold_4690 4884827 52 - 18748788 ----------------AAUUCGAUCUACAUCAUUCCAGGGUAUCA---ACAGGUGCUCA---AUUUCCACCUGU ----------------.....(((((............)).))).---(((((((....---.....))))))) ( -8.20, z-score = -0.78, R) >droYak2.chrX 7284235 52 + 21770863 ----------------AGCUUUACCUACAUGAUUCCAGGGUAUCA---GCAGGUGCUCA---ACUUGCACCUGU ----------------.(((.(((((...........)))))..)---))((((((...---....)))))).. ( -16.70, z-score = -2.13, R) >droSec1.super_21 752885 68 + 1102487 GAAUUUUUCCACUAUGAACUUAACUUGUAUUUUUCUAGGGUAUCA---ACAGGUGCUCA---AUUUCCACCUGU ........((.(((.(((..((.....))..))).))))).....---(((((((....---.....))))))) ( -10.10, z-score = -0.38, R) >droSim1.chrX 9903040 68 + 17042790 GAAUUUUUCCACUAUAAAUUUAACUUGUAUUUUUCUAGGGUAUCA---ACAGGUGCUAA---AUUUCCACCGGU ........((.(((.(((..((.....))..))).)))((((((.---...))))))..---.........)). ( -7.10, z-score = 0.67, R) >consensus G_AUUUU_CCACUAUAAACUUAACUUGCAUUAUUCUAGGGUAUCA___ACAGGUGCUCA___AUUUCCACCUGU ................................................(((((((............))))))) ( -5.11 = -5.50 + 0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:39:07 2011