| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,253,367 – 11,253,471 |
| Length | 104 |
| Max. P | 0.680584 |

| Location | 11,253,367 – 11,253,471 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 69.01 |
| Shannon entropy | 0.64090 |
| G+C content | 0.29594 |
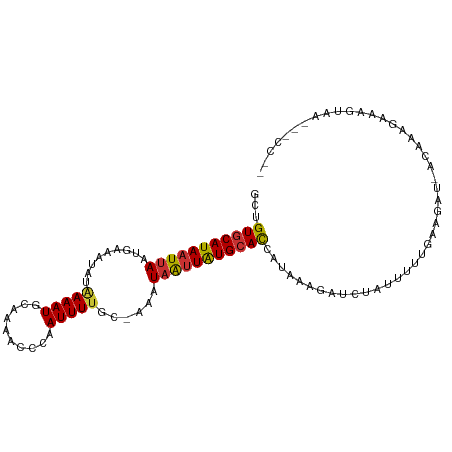
| Mean single sequence MFE | -14.99 |
| Consensus MFE | -8.35 |
| Energy contribution | -8.29 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.53 |
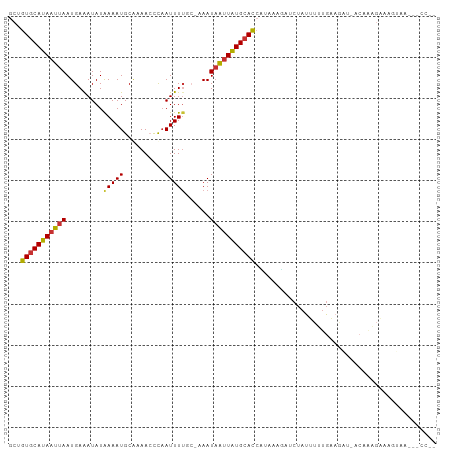
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.680584 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
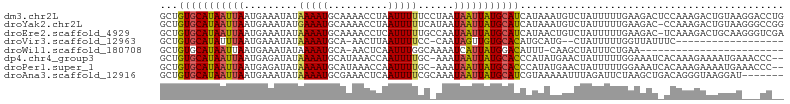
>dm3.chr2L 11253367 104 - 23011544 GCUGUGCAUAAUUAAUGAAAUAUAAAAUGCAAAACCUAAUUUUUCCUAAUAAUUAUGCAUCAUAAAUGUCUAUUUUUGAAGACUCCAAAGACUGUAAGGACCUG ...(((((((((((..((((.....................))))....))))))))))).......((((........))))(((...........))).... ( -13.20, z-score = -0.51, R) >droYak2.chr2L 7649058 103 - 22324452 GCUGUGCAUAAUUAAUGAAAUAUGAAAUGCAAAACCUAAUUUUUCAUAAUAAUUAUGCAUCAUAAAUGUCUAUUUUUGAAGAC-CCAAAGACUGUAAGGGCCGG ((((((((((((((((((((.((...............)).))))))..))))))))))).......((((........))))-..............)))... ( -17.76, z-score = -0.77, R) >droEre2.scaffold_4929 12448877 103 + 26641161 GCUGUGCAUAAUUAAUGAAAUAUAAAAUGCAAAACCUCAUUUUUGCCAAUAAUUAUGCAUCAUAACUGUCUAUUUUUGAAGAC-UCAAAGACUGCAAGGGUCGA ...(((((((((((.((...........((((((......)))))))).))))))))))).......((((........))))-.....((((.....)))).. ( -19.90, z-score = -1.22, R) >droVir3.scaffold_12963 17951185 82 + 20206255 GCUGUGCAUAUUUAAUGAAAUAUAAAAUGCA-AACUUAAUUUUCC-CAAUAGUUGUGCACAUGCAUG--CUAUUUUUGGUUAUUUC------------------ ((((((((((.(((..((((...........-........)))).-...))).)))))))).))..(--(((....))))......------------------ ( -13.21, z-score = 0.46, R) >droWil1.scaffold_180708 8057828 78 + 12563649 GCUGUGCAUAAUUAAUGAAAUAUAAAAUGCA-AACUCAAUUUGGCAAAAUCAUUAUGGACAUUU-CAAGCUAUUUCUGAA------------------------ ..(((.((((((..........((((.((..-....)).))))........)))))).)))...-...............------------------------ ( -5.87, z-score = 2.18, R) >dp4.chr4_group3 6676613 101 - 11692001 GCUGUGCAUAAUUAAUGAGAUAUAAAAUGCAUAAACCAAUUUUGC-AAAUAAUUAUGCACCCAUAUGAACUAUUUUUGGAAAUCACAAAGAAAAUGAAACCC-- ...((((((((((((((...)))....((((...........)))-)..)))))))))))............((((((.......))))))...........-- ( -15.20, z-score = -1.35, R) >droPer1.super_1 8126148 101 - 10282868 GCUGUGCAUAAUUAAUGAGAUAUAAAAUGCAUAAACCAAUUUUGC-AAAUAAUUAUGCACCCAUAUGAACUAUUUUUGGAAAUCACAAAGAAAAUGAAACCC-- ...((((((((((((((...)))....((((...........)))-)..)))))))))))............((((((.......))))))...........-- ( -15.20, z-score = -1.35, R) >droAna3.scaffold_12916 11974448 97 + 16180835 GCUGUGCAUAAUUAAUGAAAUAUAAAAUGCGAAACUCAAUUUUCGCAAAUAAUUAUGCAUCGUAAAAAUUUAGAUUCUAAGCUGACAGGGUAAGGAU------- ((.((((((((((((((...)))....(((((((......)))))))..))))))))))).))..........(((((.(.((....)).).)))))------- ( -19.60, z-score = -1.67, R) >consensus GCUGUGCAUAAUUAAUGAAAUAUAAAAUGCAAAACCCAAUUUUGC_AAAUAAUUAUGCACCAUAAAGAUCUAUUUUUGAAGAU_ACAAAGAAAGUAA___CC__ ...(((((((((((.........(((((..........)))))......)))))))))))............................................ ( -8.35 = -8.29 + -0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:56 2011