| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,927,420 – 12,927,528 |
| Length | 108 |
| Max. P | 0.649256 |

| Location | 12,927,420 – 12,927,528 |
|---|---|
| Length | 108 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 70.68 |
| Shannon entropy | 0.57303 |
| G+C content | 0.60592 |
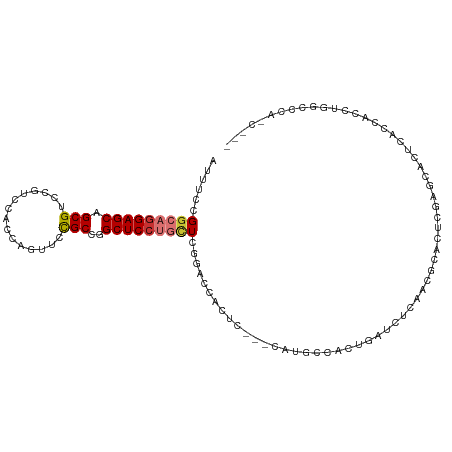
| Mean single sequence MFE | -26.01 |
| Consensus MFE | -14.11 |
| Energy contribution | -14.56 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.649256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
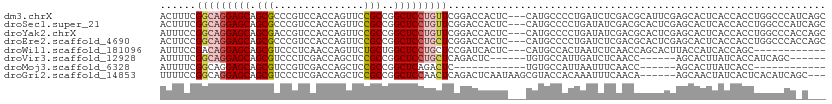
>dm3.chrX 12927420 108 + 22422827 ACUUUCGGCAGGAGCAGCGCCCGUCCACCAGUUCCGCCGGCUCCUGUUCGGACCACUC---CAUGCCCCUGAUCUCGACGCAUUCGAGCACUCACCACCUGGCCCAUCAGC ......((((.((((((.(((.((...........)).)))..))))))(((....))---).)))).((((((((((.....)))))......((....))...))))). ( -28.50, z-score = 0.28, R) >droSec1.super_21 675401 108 + 1102487 ACUUUCGGCAGGAGCAGCGCCCGUCCACCAGUUCCGCCGGCUCCUGUUCGGACCACUC---CAUGCCCCUGAUAUCGACGCACUCGAGCACUCACCACCUGGCCCAUCAGC ......((((.((((((.(((.((...........)).)))..))))))(((....))---).)))).(((((.((((.....)))).......((....))...))))). ( -27.10, z-score = 0.71, R) >droYak2.chrX 7201493 108 + 21770863 AUUUCCGGCAGGAGCAGCGACCGUCCACCAGUUCCGCCGGCUCCUGUUCGGACCACUC---CAUGCCCCUGAUAUCGACGCACUCGAGCACUCACCACCUGGCCCACCAGC ...((((((((((((.(((...............)))..))))))).)))))......---........(((..((((.....))))....)))....((((....)))). ( -29.46, z-score = -0.23, R) >droEre2.scaffold_4690 4809173 108 - 18748788 ACUUCCGGCAGGAGCAGCGCCCGUCCACCAGUUCCGCCGGCUCCUGCUCGGACCACUC---CAUGCCCCUGAUCUCGACGCACUCGAGCACUCACCACCUGGCCCACCAGC ...((((((((((((.(((..(........)...)))..)))))))).))))......---........(((.(((((.....)))))...)))....((((....)))). ( -33.90, z-score = -1.09, R) >droWil1.scaffold_181096 5696652 96 - 12416693 AUUUCCGACAGGAGCAGCGUCCCUCAACCAGUUCUGCUGGCUCCUGCUCCGAUCACUC---CAUGCCACUAAUCUCAACCAGCACUUACCAUCACCAGC------------ ......((..(((((((.(.....)..((((.....))))...)))))))..))....---......................................------------ ( -18.00, z-score = -0.80, R) >droVir3.scaffold_12928 3392609 93 - 7717345 AUUUUCGGCAGGAGCAGCGUCCCUCGACCAGCUCCGCCGGCUCCUGCUCAGACUC------UGUGCCAUUGAUCUCAACC------AGCACUUAUCACCAUCAGC------ ..(((.(((((((((.(((...((.....))...)))..))))))))).)))...------.((((..(((....)))..------.))))..............------ ( -25.50, z-score = -1.06, R) >droMoj3.scaffold_6328 298796 81 + 4453435 AUUUUCGGCAGGAGCAGCGUCCGUCGACCAGCUCCGCCGGCUCAGACUC------------UGUGCCAUUAAUUUCAACC------AGCACUUAUCACC------------ ......(((.(((((.(.(((....)))).))))))))(((.(((...)------------)).))).............------.............------------ ( -24.30, z-score = -2.71, R) >droGri2.scaffold_14853 9123062 102 - 10151454 UUUUCCGGCAGGAGCAGCGUCCCUCGACCAGCUCCGCCGGCUCCAACUCAGACUCAAUAAGCGUACCACAAAUUUCAACA------AGCAACUAUCACUCACAUCAGC--- ....(((((.(((((.(.(((....)))).))))))))))....................((..................------.))...................--- ( -21.31, z-score = -2.21, R) >consensus AUUUCCGGCAGGAGCAGCGUCCGUCCACCAGUUCCGCCGGCUCCUGCUCGGACCACUC___CAUGCCACUGAUCUCAACGCACUCGAGCACUCACCACCUGGCCCA_C___ ......(((((((((.(((...............)))..)))))))))............................................................... (-14.11 = -14.56 + 0.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:38:57 2011