| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,252,820 – 11,252,919 |
| Length | 99 |
| Max. P | 0.961510 |

| Location | 11,252,820 – 11,252,919 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 67.76 |
| Shannon entropy | 0.65735 |
| G+C content | 0.48958 |
| Mean single sequence MFE | -29.65 |
| Consensus MFE | -13.31 |
| Energy contribution | -13.02 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.961510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
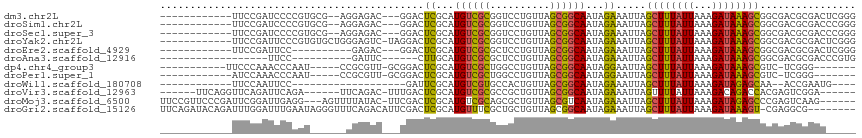
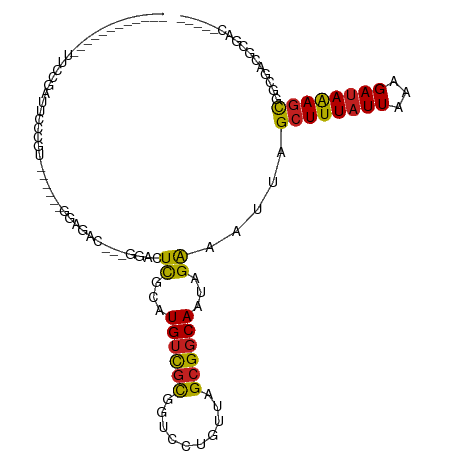
>dm3.chr2L 11252820 99 - 23011544 ------------UUCCGAUCCCCGUGCG--AGGAGAC---GGACUCGCAUGUCGCGGUCCUGUUAGCGGCAAUAGAAAUUAGCUUUAUUAAAGAUAAAGCGGCGACGCGACUCGGG ------------..((((.....(((((--((.....---...)))))))((((((((((((((......)))))......((((((((...)))))))))))..)))))))))). ( -36.30, z-score = -1.68, R) >droSim1.chr2L 11049313 99 - 22036055 ------------UUCCGAUCCCCGUGCG--AGGAGAC---GGACUCGCAUGUCGCGGUCCUGUUAGCGGCAAUAGAAAUUAGCUUUAUUAAAGAUAAAGCGGCGACGCGACCCGGG ------------........((((((((--((.....---...)))))))((((((((((((((......)))))......((((((((...)))))))))))..))))))..))) ( -35.10, z-score = -1.35, R) >droSec1.super_3 6649397 99 - 7220098 ------------UUCCGAUCCCCGUGCG--AGGAGAC---GGACUCGCAUGUCGCGGUCCUGUUAGCGGCAAUAGAAAUUAGCUUUAUUAAAGAUAAAGCGGCGACGCGACCCGGG ------------........((((((((--((.....---...)))))))((((((((((((((......)))))......((((((((...)))))))))))..))))))..))) ( -35.10, z-score = -1.35, R) >droYak2.chr2L 7648486 103 - 22324452 ------------UUCCGAUUCCCGUGUGCUGGGAGUC-UAGGACUCGCAUGUCGCGGUCCUGUUAGCGGCAAUAGAAAUUAGCUUUAUUAAAGAUAAAGCGGCGACGCGACUCGGG ------------..((((((((((.....))))))))-...((.((((..(((((....(((((......)))))......((((((((...)))))))).))))))))).)))). ( -36.80, z-score = -1.70, R) >droEre2.scaffold_4929 12448370 91 + 26641161 ------------UUCCGAUUCC----------GAGAC---GGACUCGCAUGUCGCGCUCCUGUUAGCGGCAAUAGAAAUUAGCUUUAUUAAAGAUAAAGCGGCGACGCGACUCGGG ------------........((----------(((.(---(...((((.((((((..........))))))..........((((((((...)))))))).))))..)).))))). ( -30.40, z-score = -1.45, R) >droAna3.scaffold_12916 11974069 82 + 16180835 ------------------UUCC----------GAUUC------CUUGCAUGUCGCGCUCCUGUUAGCGGCAAUAGAAAUUAGCUUUAUUAAAGAUAAAGCGGCGACGCGACCCGUG ------------------....----------.....------...((..(((((((.(((((.....)))..........((((((((...)))))))))).).))))))..)). ( -20.50, z-score = -0.28, R) >dp4.chr4_group3 6675820 91 - 11692001 -----------UUCCCAAACCCAAU-----CCGCGUU-GCGGACUCGCAUGUCGCUGGCCUGUUAGCGGCAAUAGGAAUUAGCUUUAUUAAAGAUAAAGCGUC-UCGGG------- -----------..(((....((..(-----((((...-)))))......(((((((((....)))))))))...)).....((((((((...))))))))...-..)))------- ( -30.90, z-score = -2.25, R) >droPer1.super_1 8125366 90 - 10282868 ------------AUCCAAACCCAAU-----CCGCGUU-GCGGACUCGCAUGUCGCUGGCCUGUUAGCGGCAAUAGGAAUUAGCUUUAUUAAAGAUAAAGCGUC-UCGGG------- ------------.......(((..(-----((....(-(((....))))(((((((((....)))))))))...)))....((((((((...))))))))...-..)))------- ( -30.60, z-score = -2.25, R) >droWil1.scaffold_180708 8056176 79 + 12563649 ------------UUCCAAUUCC-------------------GAUUCGCAUGUCGUGCCACUGUUAGCGGCAAUAGAAAUUAGCUUUAUUAAAGAUAGAGCAA--ACCGAAUG---- ------------..........-------------------.(((((....((.((((.(.....).))))...)).....((((((((...))))))))..--..))))).---- ( -17.30, z-score = -1.69, R) >droVir3.scaffold_12963 17950720 97 + 20206255 ------UUCAGGUUCAGAUUCAGA------UUCAGAC-UUUGACUCGCAUGUCGCGCCGCUGUUAGCGGCAAUAGAAAUUAGUUUUAUUAAAGACAGACCACGAGUCGGA------ ------...(((((..(((....)------))..)))-))(((((((..((((..(((((.....)))))(((((((.....)))))))...)))).....)))))))..------ ( -27.50, z-score = -2.14, R) >droMoj3.scaffold_6500 4882571 106 + 32352404 UUCCGUUCCCGAUUCGGAUUGAGG---AGUUUUAUAC-UUCGACUCGCAUGUCGCAGCGCUGUUAGCGUCAAUAGAAAUUAGCUUUAUUAAAGAUAGAGCCCGAGUCAAG------ ..........((((((((((((((---(((.....))-)))(((......)))....(((.....))))))))........((((((((...)))))))))))))))...------ ( -29.90, z-score = -2.08, R) >droGri2.scaffold_15126 7058522 107 + 8399593 UUCAGAUACAGAUUUGGAUUUGAAUAGGGUUUCAGACAUUCGACUCGCAUGUUUCGCUGCUGUUAGCGGCAAUAGAAAUUAGCUUUAUUAAAGAUAAAGU-CGAGGCG-------- (((((((....)))))))((((((......))))))..((((((..((..(((((..(((((....)))))...)))))..))((((((...))))))))-))))...-------- ( -25.40, z-score = -0.93, R) >consensus ____________UUCCGAUUCCCGU______GGAGAC___GGACUCGCAUGUCGCGGUCCUGUUAGCGGCAAUAGAAAUUAGCUUUAUUAAAGAUAAAGCGGCGACGCGAC_____ ............................................((...((((((..........))))))...)).....((((((((...))))))))................ (-13.31 = -13.02 + -0.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:55 2011