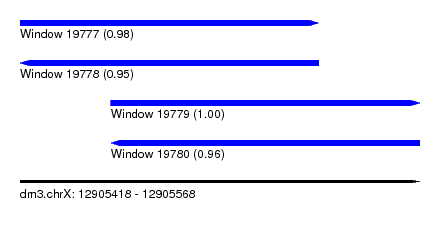
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,905,418 – 12,905,568 |
| Length | 150 |
| Max. P | 0.999193 |

| Location | 12,905,418 – 12,905,530 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.05 |
| Shannon entropy | 0.52517 |
| G+C content | 0.48450 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -19.90 |
| Energy contribution | -20.55 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.983891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12905418 112 + 22422827 ------AAUUUAAUCCAUGACAACGCAACGAUAUGCUUCUGCAAAGUCAGUCUAAUGAUGGCCACGGCCGGGGGAAACCUAUUGCCAUGGCCCACACUGUAACGGACAGCACAUGUCC-- ------...........((((...(((..((......)))))...))))(((....)))(((((.(((..(((....)))...))).)))))...........(((((.....)))))-- ( -33.90, z-score = -1.63, R) >droAna3.scaffold_13117 3834466 102 + 5790199 GAAAAAAAGGUGAUUUUUAAGGGGAUAGUUAUA-----CUAUAAUAUCAACCUAAUGAU--UUAAGGAAUGGGAACAAUUUUUGUAAACAAAAAAACUGAAAGGGAAAU----------- .......(((((((..(((.((...........-----)).))).))).))))......--........((....)).((((((....))))))..((....)).....----------- ( -10.30, z-score = 0.44, R) >droEre2.scaffold_4690 4789156 101 - 18748788 -------AUUUAAUCC-UGGCAACGCAGCGAUAUGCU-CUGCAAAGUCAGUUUGGUGAUGGUCGCGGCCGGGG-AAACCUACUGCCAUGGCCGACACUGUACCGGACGGCC--------- -------.........-.(((...((((.(.....).-))))...(((.((.(((((.((((((.(((.(((.-...)))...))).)))))).))))).))..))).)))--------- ( -37.00, z-score = -1.14, R) >droYak2.chrX 7176444 105 + 21770863 ------AAUUUAAUCCCUGGCAACGCAGCGAUAUGCCUCUGCAAAGUCAGUCUAAUGAUGGCCACGGCCUGGGGAAACCUAUUGCCAUGGCCGACACUGUAACGGACAGUG--------- ------..........(((((...((((.(......).))))...)))))........((((((.(((.((((....))))..))).)))))).((((((.....))))))--------- ( -39.90, z-score = -3.26, R) >droSec1.super_21 648804 113 + 1102487 ------AAAUUAAACCCUGACAACGCAGCGAUAUGCUUCUGCAAAGUCUGUCUAAUGAUGGCCACGGCCGGGG-AGACCUAUUGCCAUGGCCGACACUGUAACGGACAGCACAUGUCCAG ------..................((((.....(((....)))..(((.(((....)))(((((.(((.(((.-...)))...))).)))))))).))))...(((((.....))))).. ( -38.70, z-score = -2.80, R) >droSim1.chrX 9835289 113 + 17042790 ------AAUUUAAUCCCUGACAACGCAGCGAUAUGCUUCUGCAAAGUCAGUCUAAUGAUGGCCACGGCCGGGG-AGACCUAUUGCCAUGGCCGACACUGUAACGGACAGCACAUGUCCAG ------......(((.(((......))).))).(((....)))..(((.(((....)))(((((.(((.(((.-...)))...))).))))))))........(((((.....))))).. ( -39.40, z-score = -2.79, R) >consensus ______AAUUUAAUCCCUGACAACGCAGCGAUAUGCUUCUGCAAAGUCAGUCUAAUGAUGGCCACGGCCGGGG_AAACCUAUUGCCAUGGCCGACACUGUAACGGACAGCA_________ .............((((((((...((((.(......).))))...))))).....((.((((((.(((..(((....)))...))).)))))).)).......))).............. (-19.90 = -20.55 + 0.65)

| Location | 12,905,418 – 12,905,530 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.05 |
| Shannon entropy | 0.52517 |
| G+C content | 0.48450 |
| Mean single sequence MFE | -33.24 |
| Consensus MFE | -21.91 |
| Energy contribution | -24.72 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.953658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12905418 112 - 22422827 --GGACAUGUGCUGUCCGUUACAGUGUGGGCCAUGGCAAUAGGUUUCCCCCGGCCGUGGCCAUCAUUAGACUGACUUUGCAGAAGCAUAUCGUUGCGUUGUCAUGGAUUAAAUU------ --.(((((.(((.(((.(((..((((..(((((((((....((......)).)))))))))..)))).))).)))...))).).(((......)))..))))............------ ( -39.00, z-score = -1.64, R) >droAna3.scaffold_13117 3834466 102 - 5790199 -----------AUUUCCCUUUCAGUUUUUUUGUUUACAAAAAUUGUUCCCAUUCCUUAA--AUCAUUAGGUUGAUAUUAUAG-----UAUAACUAUCCCCUUAAAAAUCACCUUUUUUUC -----------..........((((((((........))))))))..............--......((((.(((.((((((-----.....)))).......)).)))))))....... ( -6.51, z-score = 0.10, R) >droEre2.scaffold_4690 4789156 101 + 18748788 ---------GGCCGUCCGGUACAGUGUCGGCCAUGGCAGUAGGUUU-CCCCGGCCGCGACCAUCACCAAACUGACUUUGCAG-AGCAUAUCGCUGCGUUGCCA-GGAUUAAAU------- ---------(((((.((......).).)))))((((..((.((((.-....))))))..)))).......(((.(..(((((-.(.....).)))))..).))-)........------- ( -28.30, z-score = 0.92, R) >droYak2.chrX 7176444 105 - 21770863 ---------CACUGUCCGUUACAGUGUCGGCCAUGGCAAUAGGUUUCCCCAGGCCGUGGCCAUCAUUAGACUGACUUUGCAGAGGCAUAUCGCUGCGUUGCCAGGGAUUAAAUU------ ---------....((((.....((((..(((((((((....((....))...)))))))))..))))...(((.(..(((((.((....)).)))))..).)))))))......------ ( -37.90, z-score = -2.03, R) >droSec1.super_21 648804 113 - 1102487 CUGGACAUGUGCUGUCCGUUACAGUGUCGGCCAUGGCAAUAGGUCU-CCCCGGCCGUGGCCAUCAUUAGACAGACUUUGCAGAAGCAUAUCGCUGCGUUGUCAGGGUUUAAUUU------ ..(((((.....)))))(((..((((..(((((((((....((...-..)).)))))))))..)))).)))(((((((((((.(((.....)))...))).)))))))).....------ ( -43.20, z-score = -2.68, R) >droSim1.chrX 9835289 113 - 17042790 CUGGACAUGUGCUGUCCGUUACAGUGUCGGCCAUGGCAAUAGGUCU-CCCCGGCCGUGGCCAUCAUUAGACUGACUUUGCAGAAGCAUAUCGCUGCGUUGUCAGGGAUUAAAUU------ ..(((((.....))))).....((((..(((((((((....((...-..)).)))))))))..))))...(((((..(((((..........)))))..)))))..........------ ( -44.50, z-score = -2.88, R) >consensus _________UGCUGUCCGUUACAGUGUCGGCCAUGGCAAUAGGUUU_CCCCGGCCGUGGCCAUCAUUAGACUGACUUUGCAGAAGCAUAUCGCUGCGUUGUCAGGGAUUAAAUU______ .............((((.....((((..(((((((((....((....))...)))))))))..))))...(((((..(((((..........)))))..)))))))))............ (-21.91 = -24.72 + 2.81)

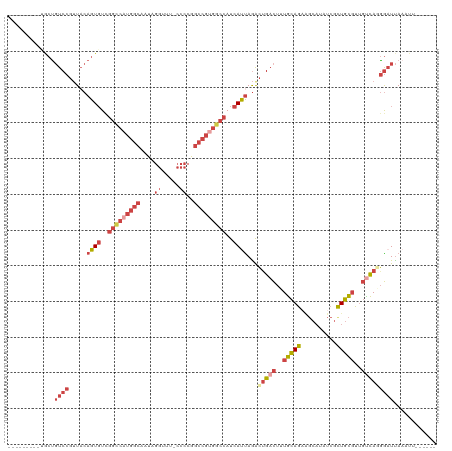
| Location | 12,905,452 – 12,905,568 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 61.90 |
| Shannon entropy | 0.70648 |
| G+C content | 0.48966 |
| Mean single sequence MFE | -35.74 |
| Consensus MFE | -15.75 |
| Energy contribution | -16.56 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
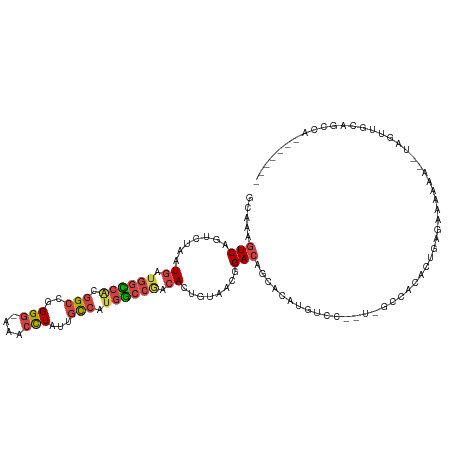
>dm3.chrX 12905452 116 + 22422827 GCAAAGUCAGUCUAAUGAUGGCCACGGCCGGGGGAAACCUAUUGCCAUGGCCCACACUGUAACGGACAGCACAUGUCC----GCCACACUGCGAAAAAAAAUAGUUGCAGCUCUCAGCUA ((...((((......))))(((((.(((..(((....)))...))).)))))....((((((((((((.....)))))----((......))...........)))))))......)).. ( -42.70, z-score = -2.98, R) >droAna3.scaffold_13117 3834501 96 + 5790199 AUAAUAUCAACCUAAUGAU--UUAAGGAAUGGGAACAAUUUUUGUAAACAAAAAAACUGAAAGGGAAAUGCAACUUUAUAAAAACACCUUGACUUCCA---------------------- .....((((......))))--((((((..((....)).((((((((((........((....))..........))))))))))..))))))......---------------------- ( -12.07, z-score = -0.18, R) >droEre2.scaffold_4690 4789187 88 - 18748788 GCAAAGUCAGUUUGGUGAUGGUCGCGGCCGGGG-AAACCUACUGCCAUGGCCGACACUGUACCGGACGGCCCUUACAU--UAGCUAUUUCA----------------------------- ((...(((.((.(((((.((((((.(((.(((.-...)))...))).)))))).))))).))..))).))........--...........----------------------------- ( -29.70, z-score = -0.84, R) >droYak2.chrX 7176478 100 + 21770863 GCAAAGUCAGUCUAAUGAUGGCCACGGCCUGGGGAAACCUAUUGCCAUGGCCGACACUGUAACGGACAGUG-----------GCCACACUGAGCGAAAA--UAGUUGUCAUCA------- .....((((......))))(((((.(((.((((....))))..))).)))))((((((((..((..(((((-----------....)))))..))...)--))).))))....------- ( -39.90, z-score = -3.48, R) >droSec1.super_21 648838 111 + 1102487 GCAAAGUCUGUCUAAUGAUGGCCACGGCCGGGG-AGACCUAUUGCCAUGGCCGACACUGUAACGGACAGCACAUGUCCAGUGACCACACUGAGAAAAAAA-UAGUUGCAGCCA------- .....(((.(((....)))(((((.(((.(((.-...)))...))).)))))))).((((((((((((.....)))))((((....))))..........-..)))))))...------- ( -44.10, z-score = -4.01, R) >droSim1.chrX 9835323 111 + 17042790 GCAAAGUCAGUCUAAUGAUGGCCACGGCCGGGG-AGACCUAUUGCCAUGGCCGACACUGUAACGGACAGCACAUGUCCAGUGGCCACACUGAGAAAAAAA-UAGUUGCAGCCA------- .....(((.(((....)))(((((.(((.(((.-...)))...))).)))))))).((((((((((((.....)))))((((....))))..........-..)))))))...------- ( -46.00, z-score = -4.12, R) >consensus GCAAAGUCAGUCUAAUGAUGGCCACGGCCGGGG_AAACCUAUUGCCAUGGCCGACACUGUAACGGACAGCACAUGUCC__U_GCCACACUGAGAAAAAA__UAGUUGCAGCCA_______ .....(((.......((.((((((.(((..(((....)))...))).)))))).))........)))..................................................... (-15.75 = -16.56 + 0.81)

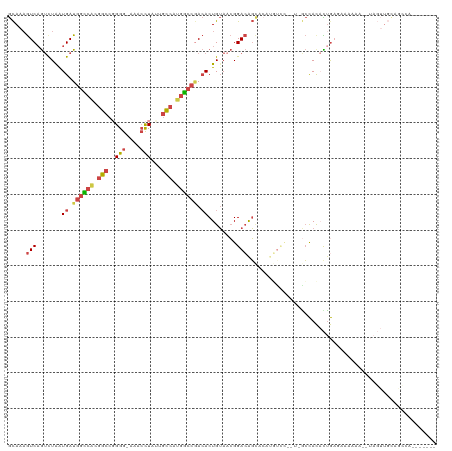
| Location | 12,905,452 – 12,905,568 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 61.90 |
| Shannon entropy | 0.70648 |
| G+C content | 0.48966 |
| Mean single sequence MFE | -32.52 |
| Consensus MFE | -16.26 |
| Energy contribution | -18.32 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
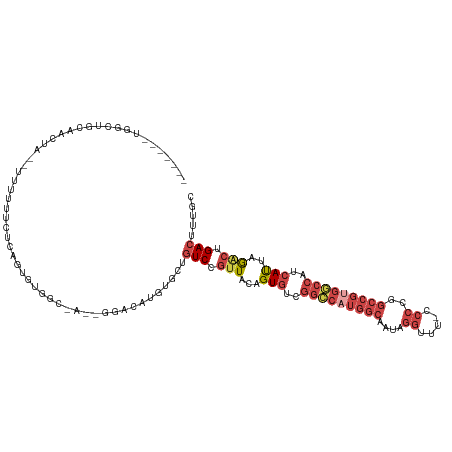
>dm3.chrX 12905452 116 - 22422827 UAGCUGAGAGCUGCAACUAUUUUUUUUCGCAGUGUGGC----GGACAUGUGCUGUCCGUUACAGUGUGGGCCAUGGCAAUAGGUUUCCCCCGGCCGUGGCCAUCAUUAGACUGACUUUGC (((((((((..................((((.((((((----(((((.....))))))))))).))))(((((((((....((......)).))))))))).)).)))).)))....... ( -48.40, z-score = -3.36, R) >droAna3.scaffold_13117 3834501 96 - 5790199 ----------------------UGGAAGUCAAGGUGUUUUUAUAAAGUUGCAUUUCCCUUUCAGUUUUUUUGUUUACAAAAAUUGUUCCCAUUCCUUAA--AUCAUUAGGUUGAUAUUAU ----------------------((((((...((((((..((....))..))))))..))))))...((((((....)))))).................--((((......))))..... ( -11.10, z-score = 0.60, R) >droEre2.scaffold_4690 4789187 88 + 18748788 -----------------------------UGAAAUAGCUA--AUGUAAGGGCCGUCCGGUACAGUGUCGGCCAUGGCAGUAGGUUU-CCCCGGCCGCGACCAUCACCAAACUGACUUUGC -----------------------------...........--..(((((((((....))).((((...((..((((..((.((((.-....))))))..))))..))..)))).)))))) ( -22.40, z-score = 1.08, R) >droYak2.chrX 7176478 100 - 21770863 -------UGAUGACAACUA--UUUUCGCUCAGUGUGGC-----------CACUGUCCGUUACAGUGUCGGCCAUGGCAAUAGGUUUCCCCAGGCCGUGGCCAUCAUUAGACUGACUUUGC -------((((((((.((.--....((..(((((....-----------)))))..))....))))))(((((((((....((....))...)))))))))))))............... ( -34.40, z-score = -1.96, R) >droSec1.super_21 648838 111 - 1102487 -------UGGCUGCAACUA-UUUUUUUCUCAGUGUGGUCACUGGACAUGUGCUGUCCGUUACAGUGUCGGCCAUGGCAAUAGGUCU-CCCCGGCCGUGGCCAUCAUUAGACAGACUUUGC -------((((..((.((.-..........))))..))))..(((((.....)))))(((..((((..(((((((((....((...-..)).)))))))))..)))).)))......... ( -38.30, z-score = -1.46, R) >droSim1.chrX 9835323 111 - 17042790 -------UGGCUGCAACUA-UUUUUUUCUCAGUGUGGCCACUGGACAUGUGCUGUCCGUUACAGUGUCGGCCAUGGCAAUAGGUCU-CCCCGGCCGUGGCCAUCAUUAGACUGACUUUGC -------((((..((.((.-..........))))..))))..(((((.....)))))(((..((((..(((((((((....((...-..)).)))))))))..)))).)))......... ( -40.50, z-score = -1.92, R) >consensus _______UGGCUGCAACUA__UUUUUUCUCAGUGUGGC_A__GGACAUGUGCUGUCCGUUACAGUGUCGGCCAUGGCAAUAGGUUU_CCCCGGCCGUGGCCAUCAUUAGACUGACUUUGC .....................................................(((.(((...(((..(((((((((....((....))...)))))))))..)))..))).)))..... (-16.26 = -18.32 + 2.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:38:52 2011