
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,893,711 – 12,893,785 |
| Length | 74 |
| Max. P | 0.518318 |

| Location | 12,893,711 – 12,893,785 |
|---|---|
| Length | 74 |
| Sequences | 9 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 74.61 |
| Shannon entropy | 0.48769 |
| G+C content | 0.48308 |
| Mean single sequence MFE | -11.57 |
| Consensus MFE | -5.69 |
| Energy contribution | -5.20 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.518318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
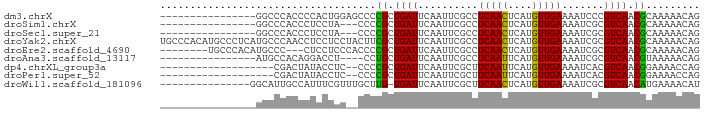
>dm3.chrX 12893711 74 + 22422827 ----------------GGCCCACCCCACUGGAGCCCCGCUGAUUCAAUUCGCCUCAACUCAUGUUGAAAAUCCCGUCAACGCAAAAACAG ----------------((((((......))).)))..((.((......))...(((((....))))).............))........ ( -13.70, z-score = -1.55, R) >droSim1.chrX 9823485 71 + 17042790 ----------------GGCCCACCCUCCUA---CCCCGCUGAUUCAAUUCGCCUCAACUCAUGUUGAAAAUCGCGUCAACGCAAAAACAG ----------------..............---.....(((............(((((....))))).....(((....))).....))) ( -9.90, z-score = -1.47, R) >droSec1.super_21 637565 71 + 1102487 ----------------GGCCCACCCUCCUA---CCCCGCUGAUUCAAUUCGCCUCAACUCAUGUUGAAAAUCGCGUCAACGCAAAAACAG ----------------..............---.....(((............(((((....))))).....(((....))).....))) ( -9.90, z-score = -1.47, R) >droYak2.chrX 7164559 90 + 21770863 UGCCCACAUGCCCUCAUGCCCAACCUCCUCCUACUUCGCUGAUUCAAUUCGCCUCAACUCAUGUUGAAAAUCGCGUCAACGCAAAAACAG ........(((....((((..................((.((......)))).(((((....))))).....))))....)))....... ( -11.40, z-score = -1.46, R) >droEre2.scaffold_4690 4778515 79 - 18748788 --------UGCCCACAUGCCC---CUCCUCCCACCCCGCUGAUUCAAUUCGCCUCAACUCAUGUUGAAAAUCGCGUCAACGCAAAAACAG --------(((....((((..---.............((.((......)))).(((((....))))).....))))....)))....... ( -11.10, z-score = -2.12, R) >droAna3.scaffold_13117 3822213 70 + 5790199 ----------------AUGCCACAGGACCU----CCUGCUGAUUCAAUUCGCCUCAAUUCAUGUUGAAAAUCGCGUCAACGUAAAAACAG ----------------....((((((....----)))).))...................(((((((........)))))))........ ( -11.50, z-score = -0.81, R) >dp4.chrXL_group3a 1180433 69 - 2690836 -------------------CGACUAUACCUC--CCCCGCUGAUUCAAUUCGCUUCAAUUCAUGUUGAAAAUCACGUCAACGGAAAACCAG -------------------............--..(((.((((...(((...((((((....)))))))))...)))).)))........ ( -9.20, z-score = -0.92, R) >droPer1.super_52 307984 69 + 549863 -------------------CGACUAUACCUC--CCCCGCUGAUUCAAUUCGCUUCAAUUCAUGUUGAAAAUCACGUCAACGGAAAACCAG -------------------............--..(((.((((...(((...((((((....)))))))))...)))).)))........ ( -9.20, z-score = -0.92, R) >droWil1.scaffold_181096 5652938 74 - 12416693 ---------------GGCAUUGCCAUUUCGUUUGCUUG-UGAUUCAAUUCGCUUCAACUCAUGUUGAAAAUCGCGUCAACAUGAAAACAU ---------------(((...))).....((((....(-(((......))))......(((((((((........))))))))))))).. ( -18.20, z-score = -1.78, R) >consensus ________________GGCCCACCCUACUUC__CCCCGCUGAUUCAAUUCGCCUCAACUCAUGUUGAAAAUCGCGUCAACGCAAAAACAG ....................................((.((((..........(((((....))))).......)))).))......... ( -5.69 = -5.20 + -0.49)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:38:47 2011