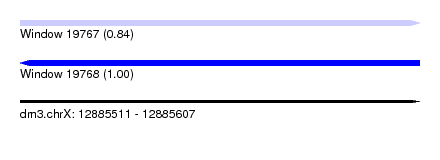
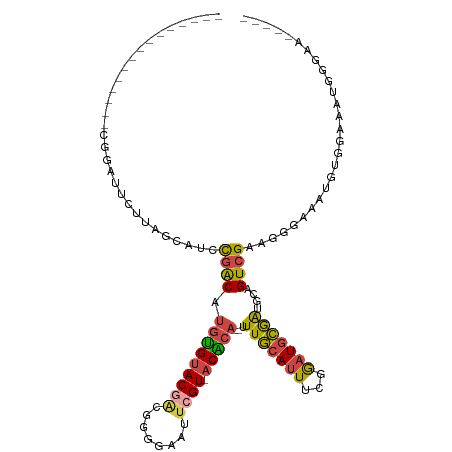
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,885,511 – 12,885,607 |
| Length | 96 |
| Max. P | 0.997036 |

| Location | 12,885,511 – 12,885,607 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 73.32 |
| Shannon entropy | 0.44980 |
| G+C content | 0.47590 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -12.88 |
| Energy contribution | -12.92 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.844691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
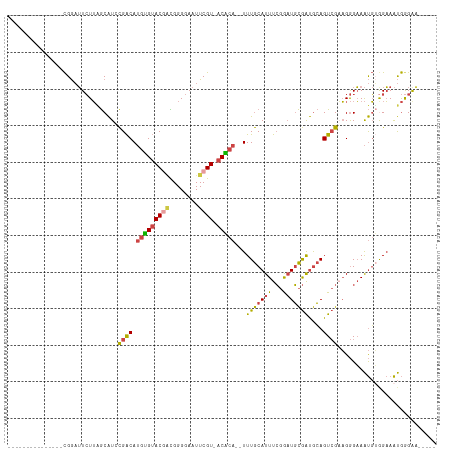
>dm3.chrX 12885511 96 + 22422827 ---------------CGGAUUCUUAGCAUCCGACAUGUGUACAGCGGGGAAUUCGU-ACACA--UUUGCAUUUCGGAUGCGAUGCAGUCGAAGGGAAAUGUGGAAAUGGGGAAA--- ---------------((((((((..((.....((....))...))..)))))))).-.((((--(((.(.(((((..(((...)))..)))))).)))))))............--- ( -27.80, z-score = -1.69, R) >droSim1.chrX 9815013 94 + 17042790 ---------------CGGAUUCUUAGCAUCCGACAUGUGUACGACGGGGAAUUCGU-ACACA--UUUGCAUUUCGGAUGCGAUGCAGUCGAAGGGAAAUGUGGAAAUGGGAA----- ---------------....((((((((((((((.((((((((((........))))-)))))--).......))))))))...(((.((....))...))).....))))))----- ( -30.70, z-score = -2.66, R) >droSec1.super_21 627789 94 + 1102487 ---------------CGGAUUCUUAGCAUCCGACAUGUGUACGACGGGGAAUUCGU-ACACA--UUUGCAUUUCGGAUGCGAUGCAGUCGAAGGGAAAUGUGGAAAUGGGAA----- ---------------....((((((((((((((.((((((((((........))))-)))))--).......))))))))...(((.((....))...))).....))))))----- ( -30.70, z-score = -2.66, R) >droYak2.chrX 7156411 96 + 21770863 ---------------CGGAUUCUUGGCAUACGACAUGUGUACGAUGGGGAAUUCGU-ACACACAUUUGCAUUUCGGAUGCGAUGCAGUCGCAGGGAAAUGUGGAAAUGGGAA----- ---------------....(((((...........(((((((((........))))-)))))..((..((((((...((((((...))))))..))))))..))...)))))----- ( -31.00, z-score = -2.98, R) >droEre2.scaffold_4690 4771666 94 - 18748788 ---------------CGGAUUCUUAGCAUGCGACAUGUGUACGAUGGGGAAUUCGU-ACGCA--UUUGCAUUUCGGAUGCGAUGCAGUCGAAGGGAAAUGGAGAAAUGAGAG----- ---------------.....(((((.(.(((((.((((((((((........))))-)))))--))))))(((((..(((...)))..))))).........)...))))).----- ( -29.00, z-score = -2.63, R) >droAna3.scaffold_13117 3816317 115 + 5790199 CAAUUCCAUUUCCCUUGAUAUGUGUACGUGUGGCAGUGGAACAUUGGGGAAUUCGUUGCCCA--UUUACAUUUGCAUUUUAGCAGAGUAGCCGAGUAGCCGAGUAGCAGGGAGAGAA ........(((((((..((.((..(((...((((.(((((....((((.((....)).))))--))))).(((((......)))))...)))).)))..)).))...)))))))... ( -31.00, z-score = -1.01, R) >consensus _______________CGGAUUCUUAGCAUCCGACAUGUGUACGACGGGGAAUUCGU_ACACA__UUUGCAUUUCGGAUGCGAUGCAGUCGAAGGGAAAUGUGGAAAUGGGAA_____ ..............................((((.(((((((((........)))).)))))...(((((((...)))))))....))))........................... (-12.88 = -12.92 + 0.03)

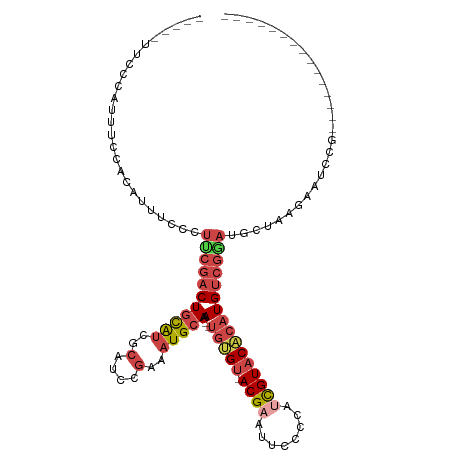
| Location | 12,885,511 – 12,885,607 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 73.32 |
| Shannon entropy | 0.44980 |
| G+C content | 0.47590 |
| Mean single sequence MFE | -24.38 |
| Consensus MFE | -10.46 |
| Energy contribution | -11.72 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.997036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12885511 96 - 22422827 ---UUUCCCCAUUUCCACAUUUCCCUUCGACUGCAUCGCAUCCGAAAUGCAAA--UGUGU-ACGAAUUCCCCGCUGUACACAUGUCGGAUGCUAAGAAUCCG--------------- ---..................................((((((((.......(--(((((-(((..........))))))))).))))))))..........--------------- ( -22.70, z-score = -3.22, R) >droSim1.chrX 9815013 94 - 17042790 -----UUCCCAUUUCCACAUUUCCCUUCGACUGCAUCGCAUCCGAAAUGCAAA--UGUGU-ACGAAUUCCCCGUCGUACACAUGUCGGAUGCUAAGAAUCCG--------------- -----................................((((((((.......(--(((((-((((........)))))))))).))))))))..........--------------- ( -26.80, z-score = -4.56, R) >droSec1.super_21 627789 94 - 1102487 -----UUCCCAUUUCCACAUUUCCCUUCGACUGCAUCGCAUCCGAAAUGCAAA--UGUGU-ACGAAUUCCCCGUCGUACACAUGUCGGAUGCUAAGAAUCCG--------------- -----................................((((((((.......(--(((((-((((........)))))))))).))))))))..........--------------- ( -26.80, z-score = -4.56, R) >droYak2.chrX 7156411 96 - 21770863 -----UUCCCAUUUCCACAUUUCCCUGCGACUGCAUCGCAUCCGAAAUGCAAAUGUGUGU-ACGAAUUCCCCAUCGUACACAUGUCGUAUGCCAAGAAUCCG--------------- -----....................(((((((((((..(....)..)))))...((((((-((((........)))))))))))))))).............--------------- ( -25.40, z-score = -4.33, R) >droEre2.scaffold_4690 4771666 94 + 18748788 -----CUCUCAUUUCUCCAUUUCCCUUCGACUGCAUCGCAUCCGAAAUGCAAA--UGCGU-ACGAAUUCCCCAUCGUACACAUGUCGCAUGCUAAGAAUCCG--------------- -----.......((((.........((((..(((...)))..))))((((..(--((.((-((((........)))))).)))...))))....))))....--------------- ( -19.40, z-score = -2.99, R) >droAna3.scaffold_13117 3816317 115 - 5790199 UUCUCUCCCUGCUACUCGGCUACUCGGCUACUCUGCUAAAAUGCAAAUGUAAA--UGGGCAACGAAUUCCCCAAUGUUCCACUGCCACACGUACACAUAUCAAGGGAAAUGGAAUUG ((((.(((((.......((((....))))....(((......)))..((((..--((((((..((((........))))...)))).))..)))).......)))))...))))... ( -25.20, z-score = -1.45, R) >consensus _____UUCCCAUUUCCACAUUUCCCUUCGACUGCAUCGCAUCCGAAAUGCAAA__UGUGU_ACGAAUUCCCCAUCGUACACAUGUCGGAUGCUAAGAAUCCG_______________ .........................(((((((((((..(....)..)))))....(((((.((((........))))))))).))))))............................ (-10.46 = -11.72 + 1.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:38:42 2011