
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,885,318 – 12,885,396 |
| Length | 78 |
| Max. P | 0.710685 |

| Location | 12,885,318 – 12,885,396 |
|---|---|
| Length | 78 |
| Sequences | 9 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 84.87 |
| Shannon entropy | 0.30931 |
| G+C content | 0.47255 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -14.87 |
| Energy contribution | -14.87 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.710685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12885318 78 + 22422827 UGCUCAUUAGUAGCACAUUUACCGAGUACGUGAUUUAGCUGCAAAUGUGCAAAGGACCGUCGUCGGUCCGAAUGACUG (((.((((.(((((....((((.......))))....))))).)))).)))..((((((....))))))......... ( -26.70, z-score = -3.08, R) >droSim1.chrX 9814829 75 + 17042790 CGCUCAUUAGUAGCACAUUUACCGAGUACGUGAUUUAGCUGCAAAUGUGCAAAG---GACCGUCGGUCCGAAUGACUG .((.((((.(((((....((((.......))))....))))).)))).))...(---((((...)))))......... ( -22.30, z-score = -2.06, R) >droSec1.super_21 627605 75 + 1102487 CGCUCAUUAGUAGCACAUUUACCGAGUACGUGAUUUAGCUGCAAAUGUGCAAAG---GACCGUCGGUCCGAAUGACUG .((.((((.(((((....((((.......))))....))))).)))).))...(---((((...)))))......... ( -22.30, z-score = -2.06, R) >droYak2.chrX 7156186 75 + 21770863 CGCUCAUUAGUAGCACAUUUACCGAGUACGUGAUUUAGCUGCAAAUGUGCAAAG---GACCGUCGGUCCGAAUGACUG .((.((((.(((((....((((.......))))....))))).)))).))...(---((((...)))))......... ( -22.30, z-score = -2.06, R) >droEre2.scaffold_4690 4771486 75 - 18748788 CGCUCAUUAGUAGCACAUUUACCGAGUACGUGAUUCAGCUGCAAAUGUGCAAAG---GACCGUCGGUCCGAGUGACUG (((((.......((((((((.....(((.((......)))))))))))))...(---((((...)))))))))).... ( -24.10, z-score = -2.08, R) >droAna3.scaffold_13117 3816112 71 + 5790199 CGCUCAUUAGUAGCACAUUUACCGAGUACGUGAUUUAGCUGCAAAUGUGCAAAAAA-GAGCGACGGACCGAC------ (((((.......((((((((.....(((.((......)))))))))))))......-)))))..........------ ( -16.02, z-score = -0.89, R) >dp4.chrXL_group3a 1172854 66 - 2690836 CGCUCAUUAGUAGCACAUUUACCGAGUACGUGAUUUAGCUGCAAAUGUGCAAAG---GACCGUGCGUCG--------- .((.((((.(((((....((((.......))))....))))).)))).))....---(((.....))).--------- ( -16.50, z-score = -0.52, R) >droPer1.super_52 300033 66 + 549863 CGCUCAUUAGUAGCACAUUUACCGAGUACGUGAUUUAGCUGCAAAUGUGCAAAG---GACCGUGCGUCG--------- .((.((((.(((((....((((.......))))....))))).)))).))....---(((.....))).--------- ( -16.50, z-score = -0.52, R) >droWil1.scaffold_181096 5645177 78 - 12416693 CGCUCAUUAGUAGCACAUUUACCGAGUACGUGAUUUAGCUGCAAAUGUGCAAAAGACUGUGCAUAUUCCAUAUGUAUG .((.((((.(((((....((((.......))))....))))).)))).))........((((((((...)))))))). ( -19.60, z-score = -1.36, R) >consensus CGCUCAUUAGUAGCACAUUUACCGAGUACGUGAUUUAGCUGCAAAUGUGCAAAG___GACCGUCGGUCCGAAUGACUG .((.((((.(((((....((((.......))))....))))).)))).))............................ (-14.87 = -14.87 + -0.00)

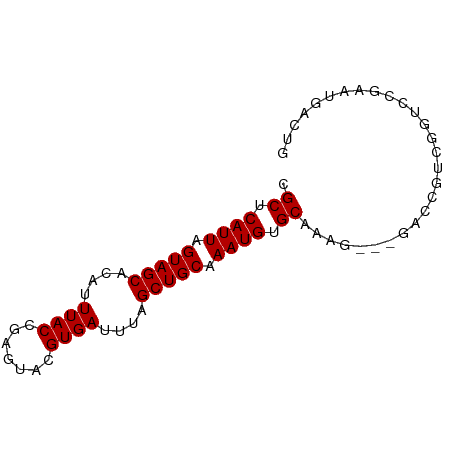
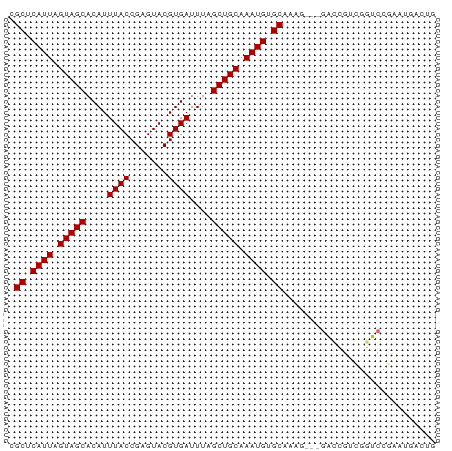
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:38:40 2011