| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,884,100 – 12,884,199 |
| Length | 99 |
| Max. P | 0.971688 |

| Location | 12,884,100 – 12,884,199 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 69.21 |
| Shannon entropy | 0.50206 |
| G+C content | 0.36090 |
| Mean single sequence MFE | -22.78 |
| Consensus MFE | -11.55 |
| Energy contribution | -11.17 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.971688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
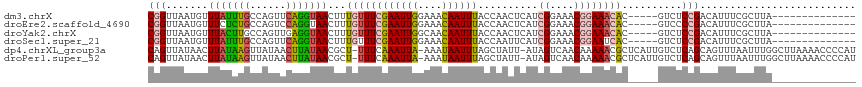
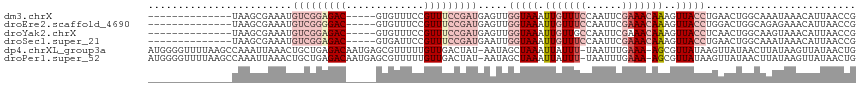
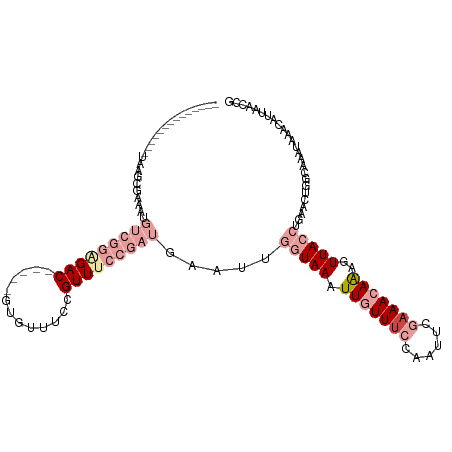
>dm3.chrX 12884100 99 + 22422827 CGGUUAAUGUUUAUUUGCCAGUUCAGGUAACUUUGUUUCGAAUUGGAAACAAUUUACCAACUCAUCGGAAACGGAAACAC-----GUCUCCGACAUUUCGCUUA-------------- .(((.(((....))).)))......(((((..(((((((......))))))).)))))......(((((.(((......)-----)).)))))...........-------------- ( -25.80, z-score = -2.50, R) >droEre2.scaffold_4690 4770361 99 - 18748788 CGGUUAAUGUUUCUCUGCCAGUCCAGGUAACUUUGUUUCGAAUUGGAAACAAUUUACCAACUCAUCGGAAACGGAAACAC-----GUCCCCGACAUUUCGCUUA-------------- (((....((((((............(((((..(((((((......))))))).))))).......((....)))))))).-----....)))............-------------- ( -24.30, z-score = -1.56, R) >droYak2.chrX 7154940 99 + 21770863 CGGUUAAUGUUUACUUGCCAGUUGAGGUAACUUUGUUUCGAAUUGGCAACAAUUUACCAACUCAUCGGAAACGGAAACAC-----GUCUCCGACAUUUCGCUUA-------------- .(((.(((......(((((((((((..((....))..)).)))))))))..))).)))......(((((.(((......)-----)).)))))...........-------------- ( -26.00, z-score = -2.24, R) >droSec1.super_21 626439 99 + 1102487 CGGUUAAUGUUUAUUUGCCAGUUCAGGUAACUUUGUUUCGAAUUGGAAACAAUUUACCAAUUCAUCGGAAACGGAAUCAC-----GUCUCCGACAUUUCGCUUA-------------- .(((.(((....))).)))......(((((..(((((((......))))))).)))))......(((((.(((......)-----)).)))))...........-------------- ( -25.80, z-score = -2.59, R) >dp4.chrXL_group3a 1171718 115 - 2690836 CAGUUAUAACUUAUAAGUUAUAACUUAUAACGCU-UUUCAAAUUA-AAAUAAUUUAGCUAUU-AUAGUCAACAAAAACGCUCAUUGUCUCAGCAGUUUAAUUUGGCUUAAAACCCCAU .((((((((((....))))))))))......(((-....((((((-...)))))))))..((-(.((((((...(((((((.........))).))))...)))))))))........ ( -17.40, z-score = -2.00, R) >droPer1.super_52 298896 115 + 549863 CAGUUAUAACUUAUAAGUUAUAACUUAUAACGCU-UUUCAAAUUA-AAAUAAUUUAGCUAUU-AUAGUCAACAAAAACGCUCAUUGUCUCAGCAGUUUAAUUUGGCUUAAAACCCCAU .((((((((((....))))))))))......(((-....((((((-...)))))))))..((-(.((((((...(((((((.........))).))))...)))))))))........ ( -17.40, z-score = -2.00, R) >consensus CGGUUAAUGUUUAUUUGCCAGUUCAGGUAACUUUGUUUCGAAUUGGAAACAAUUUACCAACUCAUCGGAAACGGAAACAC_____GUCUCCGACAUUUCGCUUA______________ (((.......(((((((......)))))))...((((((((((((....))))))..........((....))))))))..........))).......................... (-11.55 = -11.17 + -0.38)
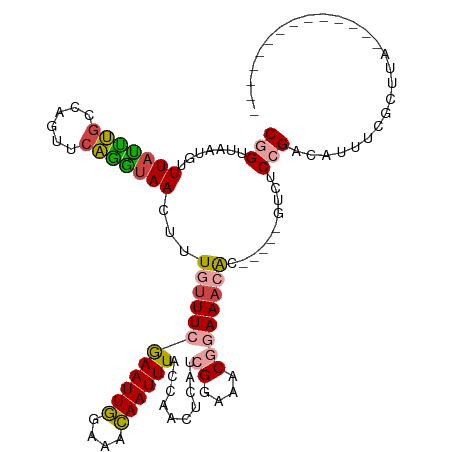
| Location | 12,884,100 – 12,884,199 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 69.21 |
| Shannon entropy | 0.50206 |
| G+C content | 0.36090 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -8.48 |
| Energy contribution | -8.70 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.735957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12884100 99 - 22422827 --------------UAAGCGAAAUGUCGGAGAC-----GUGUUUCCGUUUCCGAUGAGUUGGUAAAUUGUUUCCAAUUCGAAACAAAGUUACCUGAACUGGCAAAUAAACAUUAACCG --------------...((.....(((((((((-----(......)))))))))).(((((((((.(((((((......)))))))..)))))..)))).))................ ( -28.80, z-score = -3.38, R) >droEre2.scaffold_4690 4770361 99 + 18748788 --------------UAAGCGAAAUGUCGGGGAC-----GUGUUUCCGUUUCCGAUGAGUUGGUAAAUUGUUUCCAAUUCGAAACAAAGUUACCUGGACUGGCAGAGAAACAUUAACCG --------------...((.....(((((..((-----(......)))..))))).(((((((((.(((((((......)))))))..)))))..)))).))................ ( -28.40, z-score = -1.84, R) >droYak2.chrX 7154940 99 - 21770863 --------------UAAGCGAAAUGUCGGAGAC-----GUGUUUCCGUUUCCGAUGAGUUGGUAAAUUGUUGCCAAUUCGAAACAAAGUUACCUCAACUGGCAAGUAAACAUUAACCG --------------...((.....(((((((((-----(......))))))))))((((((((((....)))))))))).....................))................ ( -28.30, z-score = -2.89, R) >droSec1.super_21 626439 99 - 1102487 --------------UAAGCGAAAUGUCGGAGAC-----GUGAUUCCGUUUCCGAUGAAUUGGUAAAUUGUUUCCAAUUCGAAACAAAGUUACCUGAACUGGCAAAUAAACAUUAACCG --------------...((.....(((((((((-----(......)))))))))).....(((((.(((((((......)))))))..))))).......))................ ( -27.50, z-score = -3.34, R) >dp4.chrXL_group3a 1171718 115 + 2690836 AUGGGGUUUUAAGCCAAAUUAAACUGCUGAGACAAUGAGCGUUUUUGUUGACUAU-AAUAGCUAAAUUAUUU-UAAUUUGAAA-AGCGUUAUAAGUUAUAACUUAUAAGUUAUAACUG ((((.(((((....(((((((((..(((.(.....).)))(((.((((.....))-)).)))........))-))))))).))-))).)))).((((((((((....)))))))))). ( -23.30, z-score = -1.57, R) >droPer1.super_52 298896 115 - 549863 AUGGGGUUUUAAGCCAAAUUAAACUGCUGAGACAAUGAGCGUUUUUGUUGACUAU-AAUAGCUAAAUUAUUU-UAAUUUGAAA-AGCGUUAUAAGUUAUAACUUAUAAGUUAUAACUG ((((.(((((....(((((((((..(((.(.....).)))(((.((((.....))-)).)))........))-))))))).))-))).)))).((((((((((....)))))))))). ( -23.30, z-score = -1.57, R) >consensus ______________UAAGCGAAAUGUCGGAGAC_____GUGUUUCCGUUUCCGAUGAAUUGGUAAAUUGUUUCCAAUUCGAAACAAAGUUACCUGAACUGGCAAAUAAACAUUAACCG ..........................(((((((.......))))))).......((((((((..........)))))))).......((((.......))))................ ( -8.48 = -8.70 + 0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:38:40 2011