| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,869,334 – 12,869,424 |
| Length | 90 |
| Max. P | 0.928708 |

| Location | 12,869,334 – 12,869,424 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 60.03 |
| Shannon entropy | 0.77200 |
| G+C content | 0.52463 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -11.16 |
| Energy contribution | -10.34 |
| Covariance contribution | -0.82 |
| Combinations/Pair | 2.08 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12869334 90 + 22422827 ---AAGGGCCUGGGCGUGGUGCUGCUGCUCCUAGGAGCAGGAGUGGCCAGCAUUAUGUAUACUUUCUUCGGAAUGGUAGCAAACUUUGAAAUU------ ---(((.((....((((((((((((..(((((......)))))..).))))))))))).(((((((....))).))))))...))).......------ ( -34.70, z-score = -2.30, R) >droSim1.chrX 9799589 90 + 17042790 ---AAGGUCCUGGGCGUGGUGGUGCUGCUCCUAGGUGCAGGAGUGGCCAGCAUUACGUAUACUUUUUUCGGAAUCGGAGCAAACUUUGAAAUG------ ---(((((.....((((((((.(((..(((((......)))))..).)).))))))))..)))))........((((((....))))))....------ ( -29.20, z-score = -1.03, R) >droSec1.super_21 611707 90 + 1102487 ---GAGGUCCUGGGCGUGGUGGUGCUGCUCCGAGGAGCAGGAGUGGCCAGCAUUACGUAUACUUUCUUUGGAACGGUGGCGAACUUUGAAAUG------ ---...((((((.((((((((.(((..((((........))))..).)).)))))))).....(((....)))))).))).............------ ( -29.80, z-score = -0.69, R) >droYak2.chrX 7141267 93 + 21770863 GCGGUCCUCCUGGGUGUGGUGCUGCUGCGCCGAGGAGCAGGAGUGGCCAGCAUUACGUAUACCUUCUUCGGGAUGGUGGCGAACUUGGAAAUG------ ((((((((((((....((((((....)))))).....)))))).)))).))....(((.(((((((....))).)))))))............------ ( -38.40, z-score = -1.65, R) >droEre2.scaffold_4690 4755534 90 - 18748788 ---GAGGGCCUGGGUGUGGUGCUGCUGCUCCAAGGAGCAGGAGUGGCCAGCAUUAUGUAUACUUUCUUCGGGUUGGUGGCAAACUUGGCAUUG------ ---....(((.(((..(((((((((..((((........))))..).))))))))..).((((.((....))..)))).....)).)))....------ ( -33.50, z-score = -0.93, R) >droAna3.scaffold_13117 3798498 77 + 5790199 ---GUGCCAUGGGCUCUAAAGGGCUUGCUUUUUGGGGUGGUGGUGGCGAGGAGAAUAUAUAUCUUCUGAGGUUUCUCCUU------------------- ---..(((((.((((((((((((....)))))))))))..).)))))((((((((.((...((....)).))))))))))------------------- ( -29.20, z-score = -2.87, R) >droWil1.scaffold_181096 11296147 99 + 12416693 ACCGUGGUGCUGGCACUGGUGGUGUUAGCACUAGUGGUGCUUGAAUUGAUUGCAGCUUUCAUUCUGUCCGCUCUGGUUGUUAUCAGAAAAUGUACAUUA ..((((((((((((((.....)))))))))))).))((((.....(((((.((((((.................)))))).))))).....)))).... ( -30.93, z-score = -1.43, R) >consensus ___GAGGUCCUGGGCGUGGUGGUGCUGCUCCUAGGAGCAGGAGUGGCCAGCAUUACGUAUACUUUCUUCGGAAUGGUGGCAAACUUUGAAAUG______ .....(.((((.((.((((.....)))).)).)))).)..((((.((((.((((.((...........)).)))).))))..))))............. (-11.16 = -10.34 + -0.82)

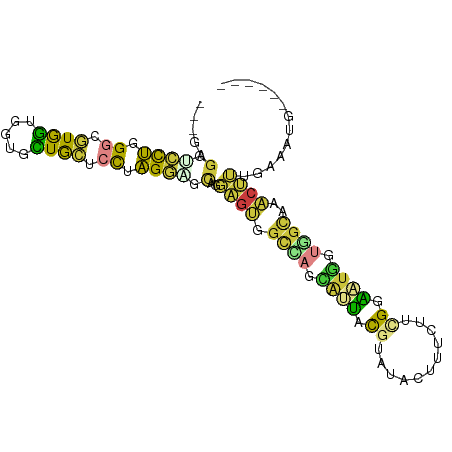
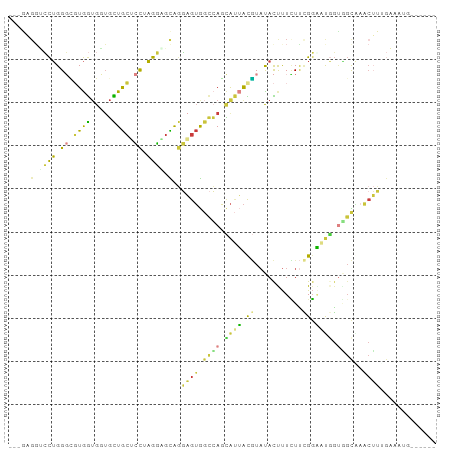
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:38:37 2011