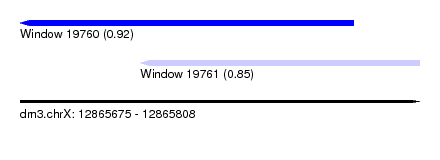
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,865,675 – 12,865,808 |
| Length | 133 |
| Max. P | 0.920550 |

| Location | 12,865,675 – 12,865,786 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 88.59 |
| Shannon entropy | 0.15719 |
| G+C content | 0.47748 |
| Mean single sequence MFE | -39.47 |
| Consensus MFE | -33.91 |
| Energy contribution | -35.47 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
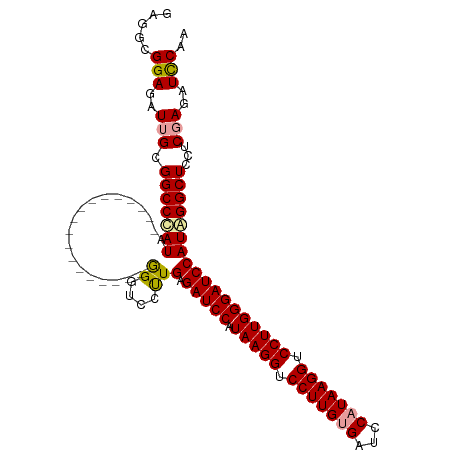
>dm3.chrX 12865675 111 - 22422827 GGGUCCUUGAGAUCCAUAAGGUCCUUGUGAUCCAUAAGGUCCUUGGGAUCCAUAGGCUCCACGAGAUCCAACGUUCGGUUGGUAAGGAUUAAUCGCAUAGGGAUUGUAGUU ((..(((((.(((((.(((((.(((((((...))))))).)))))))))))).)))..))((..(((((...((.((((((((....)))))))).))..)))))...)). ( -42.10, z-score = -2.90, R) >droSec1.super_21 607971 111 - 1102487 AGGUCCUUGAGAUCCAUAAGGUCCUUGUGAUUCAUAAGGUCCUUGGGAUCCAUGGGCUCCUCGAGAUCCAACGUUCGGUUGGUAAGGAUUAAUCGCAUAGGGAUUGUAGUU .(((((.((.(((((.(((((.(((((((...))))))).)))))))))))).)))))......(((((...((.((((((((....)))))))).))..)))))...... ( -41.50, z-score = -2.78, R) >droEre2.scaffold_4690 4751393 111 + 18748788 GGAACUCUGAGAUCCGUAAGGUCCUUGAGACCCGUAAGGUCCUUGGGAUCCAUAGGCUCUUCCAGUUUCGAUGCUCGGUUGGUAAGGAUUAAUCGCAUAGGGAUUGUAGUU ((..((.((.(((((.(((((.(((((.......))))).)))))))))))).))..))...((((..(.((((..(((((((....))))))))))).)..))))..... ( -34.80, z-score = -0.59, R) >consensus GGGUCCUUGAGAUCCAUAAGGUCCUUGUGAUCCAUAAGGUCCUUGGGAUCCAUAGGCUCCUCGAGAUCCAACGUUCGGUUGGUAAGGAUUAAUCGCAUAGGGAUUGUAGUU ((((((.((.(((((.(((((.(((((((...))))))).)))))))))))).)))))).....(((((......((((((((....)))))))).....)))))...... (-33.91 = -35.47 + 1.56)


| Location | 12,865,715 – 12,865,808 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.51 |
| Shannon entropy | 0.30610 |
| G+C content | 0.53502 |
| Mean single sequence MFE | -39.04 |
| Consensus MFE | -30.68 |
| Energy contribution | -30.80 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.850758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12865715 93 - 22422827 GAGGCGGAGAUUGCGGCCCAUA------------------GGGUCCUUGAGAUCCAUAAGGUCCUUGUGAUCCAUAAGGUCCUUGGGAUCCAUAGGCUCCACGAGAUCCAA .....(((..(((.(((((...------------------))).(((((.(((((.(((((.(((((((...))))))).)))))))))))).)))..)).)))..))).. ( -36.60, z-score = -1.81, R) >droSec1.super_21 608011 93 - 1102487 GAGGCGGAGAUUGCGGCCCAUA------------------AGGUCCUUGAGAUCCAUAAGGUCCUUGUGAUUCAUAAGGUCCUUGGGAUCCAUGGGCUCCUCGAGAUCCAA .....(((..(((.((......------------------.(((((.((.(((((.(((((.(((((((...))))))).)))))))))))).))))))).)))..))).. ( -38.81, z-score = -2.36, R) >droEre2.scaffold_4690 4751433 111 + 18748788 GAGGGGGAGAUUGCGGCCUAUAUGAACCGUAGGCUUCAGGGGAACUCUGAGAUCCGUAAGGUCCUUGAGACCCGUAAGGUCCUUGGGAUCCAUAGGCUCUUCCAGUUUCGA ..(((((((.(((.(((((((.......))))))).))).....((.((.(((((.(((((.(((((.......))))).)))))))))))).)).)))))))........ ( -41.70, z-score = -0.70, R) >consensus GAGGCGGAGAUUGCGGCCCAUA__________________GGGUCCUUGAGAUCCAUAAGGUCCUUGUGAUCCAUAAGGUCCUUGGGAUCCAUAGGCUCCUCGAGAUCCAA .....(((..(((.(((((((...................((....))(.(((((.(((((.(((((((...))))))).))))))))))))))))))...)))..))).. (-30.68 = -30.80 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:38:36 2011