| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,842,210 – 12,842,283 |
| Length | 73 |
| Max. P | 0.771549 |

| Location | 12,842,210 – 12,842,283 |
|---|---|
| Length | 73 |
| Sequences | 9 |
| Columns | 86 |
| Reading direction | reverse |
| Mean pairwise identity | 67.53 |
| Shannon entropy | 0.61747 |
| G+C content | 0.45431 |
| Mean single sequence MFE | -16.38 |
| Consensus MFE | -8.40 |
| Energy contribution | -8.62 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.771549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
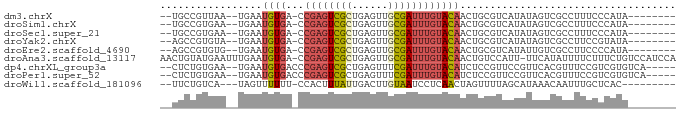
>dm3.chrX 12842210 73 - 22422827 --UGCCGUUAA--UGAAUGUGA-CCGAGUCGCUGAGUUGCGAUUUGUACAACUGCGUCAUAUAGUCGCCUUUCCCAUA-------- --........(--((..((((.-.((((((((......))))))))))))...(((.(.....).)))......))).-------- ( -15.80, z-score = -0.70, R) >droSim1.chrX 9778650 73 - 17042790 --UGCCGUGAA--UGAAUGUGA-CCGAGUCGCUGAGUUGCGAUUUGUACAACUGCGUCAUAUAGUCGCCUUUCCCAUA-------- --....((((.--...((((((-(.(((((((......)))))))(((....))))))))))..))))..........-------- ( -19.20, z-score = -1.74, R) >droSec1.super_21 584424 73 - 1102487 --UGCCGUGAA--UGAAUGUGA-CCGAGUCGCUGAGUUGCGAUUUGUACAACUGCGUCAUAUAGUCGCCUUUCCCAUA-------- --....((((.--...((((((-(.(((((((......)))))))(((....))))))))))..))))..........-------- ( -19.20, z-score = -1.74, R) >droYak2.chrX 7112070 73 - 21770863 --AGCCGUGUA--UGAAUGUGA-CCGAGUCGCUGAGUUGCGAUUUGUACAACUGCGUCAUAUAGUCGCCUUCCGUAUA-------- --.((..((((--(((...((.-.((((((((......))))))))..))......)))))))...))..........-------- ( -18.40, z-score = -0.77, R) >droEre2.scaffold_4690 4729300 73 + 18748788 --AGCCGUGUG--UGAAUGUGA-CCGAGUCGCUGAGUUGCGAUUUGUACAACUGCGUCAUAUUGUCGCCUUCCCCAUA-------- --....(.(((--..(((((((-(.(((((((......)))))))(((....)))))))))))..)))).........-------- ( -19.40, z-score = -1.63, R) >droAna3.scaffold_13117 3771168 84 - 5790199 AACUGUAUGAAUUUGAAUGUGA-CCGAGUCGCUGAGUUGCGAUUUGUACAACUGUCCAUU-UUCAUAUUUUCUUUCUGUCCAUCCA ....(((((((..((.((.((.-.((((((((......))))))))..))...)).))..-))))))).................. ( -18.60, z-score = -2.20, R) >dp4.chrXL_group3a 1103694 77 + 2690836 --CUCUGUGAA--UGAAUGUGACCCGAGUCGCUGAGUUUCGAUUUGUACAUCUCCGUUCCGUUCACGUUUCCGUCGUGUCA----- --....(((((--((((((.((..(((((((........)))))))....))..)))).)))))))...............----- ( -15.80, z-score = -0.05, R) >droPer1.super_52 232283 77 - 549863 --CUCUGUGAA--UGAAUGUGACCCGAGUCGCUGAGUUUCGAUUUGUACAUCUCCGUUCCGUUCACGUUUCCGUCGUGUCA----- --....(((((--((((((.((..(((((((........)))))))....))..)))).)))))))...............----- ( -15.80, z-score = -0.05, R) >droWil1.scaffold_181096 4987530 71 - 12416693 --UUCUGUCA---UAGUUUUUU-CCACUUUAUUGACUUGUAAUCCUCAACUAGUUUUAGCAUAAACAAUUUGCUCAC--------- --....((((---.(((.....-..)))....)))).....................((((.........))))...--------- ( -5.20, z-score = 0.21, R) >consensus __UGCCGUGAA__UGAAUGUGA_CCGAGUCGCUGAGUUGCGAUUUGUACAACUGCGUCAUAUAGUCGCCUUCCCCAUA________ .................((((...((((((((......)))))))))))).................................... ( -8.40 = -8.62 + 0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:38:31 2011