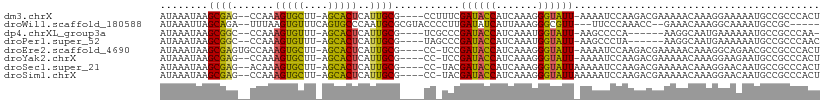
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,766,057 – 12,766,159 |
| Length | 102 |
| Max. P | 0.683147 |

| Location | 12,766,057 – 12,766,159 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.11 |
| Shannon entropy | 0.40460 |
| G+C content | 0.43197 |
| Mean single sequence MFE | -30.41 |
| Consensus MFE | -12.02 |
| Energy contribution | -11.85 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.575838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
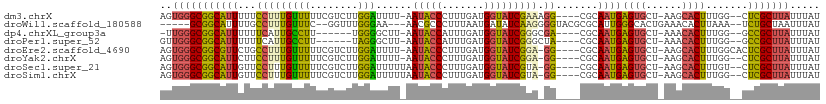
>dm3.chrX 12766057 102 + 22422827 AGUGGGCGGCAUUUUUCCUUUGUUUUUCGUCUUGGAUUUU-AAUACCCUUUGAUGGUAUCGAAAGG----CGCAAUGAGUGCU-AAGCACUUUGG--CUCGCUUAUUUAU (((((((((............((((((((...........-.(((((.......))))))))))))----)((...((((((.-..))))))..)--))))))))))... ( -26.20, z-score = -0.91, R) >droWil1.scaffold_180588 858445 98 + 1294757 -----GCGGCAUUUUGCCUUUGUUUC--GGUUUGGGAA---AACGCCCUUUAAUGAUAUCAAGGGGUACGCGCAUUGGGCACUGAAACACUUAAA--UCUGCUAAUUUAU -----(((((.....)))..((((((--(((.......---...(((((((.........)))))))..((.(...).)))))))))))......--...))........ ( -24.30, z-score = -0.75, R) >dp4.chrXL_group3a 1009780 95 - 2690836 -UUGGGCGGCAUUUUUUCAUUGCCUU------UGGGGCUU-AAUACCAUUUGAUGGUAUCGGGCGA----CGCAAUGAGUGCU-AAACACUUUGG--GCCGCUUAUUUAU -.((((((((......(((((((...------.(..(((.-.(((((((...)))))))..)))..----))))))))(((..-...))).....--))))))))..... ( -35.40, z-score = -4.00, R) >droPer1.super_52 134598 96 + 549863 GUUGGGCGGCAUUUUUUCAUUGCCUU------UAGGGCUU-AAUACCAUUUGAUGGUAUCGGGCUA----CGCAAUGAGUGCU-AAACACUUUGG--GCCGCUUAUUUAU ..((((((((......(((((((...------...((((.-.(((((((...)))))))..)))).----.)))))))(((..-...))).....--))))))))..... ( -36.30, z-score = -4.42, R) >droEre2.scaffold_4690 4661630 103 - 18748788 AGUGGGCGGCGUUCUGCCUUUGUUUUUCGUCUUGGAUUUU-AAUACCCUUUGAUGGUAUCGGA-GG----CGCAAUGAGUGCU-AAGCACUUUGGCACUCGCUUAUUUAU (((((((...(((.((((((((....(((((..((.....-.....))...)))))...))))-))----)).)))(((((((-(((...)))))))))))))))))... ( -34.90, z-score = -2.73, R) >droYak2.chrX 7042260 101 + 21770863 AGUGGGCGGCAUUCUUCCUUUGUUUUUCGUCUUGGAUUUU-AAUACCCUUUGAUGGUAUCGGA-GG----CGCAAUGAGUGCU-AAGCACUUUGG--CUCGCUUAUUUAU (((((((((.......((((((....(((((..((.....-.....))...)))))...))))-))----.((...((((((.-..))))))..)--))))))))))... ( -28.30, z-score = -1.30, R) >droSec1.super_21 516141 102 + 1102487 AGUGGGCGGCAUUGUUCCUUUGUUUUUCGUCUUGGAUUUUUAAUACCCUUUGAUGGUAUCGUA-GG----CGCAAUGAGUGCU-AAGCACUUUGU--CUCGCUUAUUUAU (((((((((((((((.(((.((....(((((..((...........))...)))))...)).)-))----.))))))(((((.-..)))))....--.)))))))))... ( -28.10, z-score = -2.05, R) >droSim1.chrX 9734191 102 + 17042790 AGUGGGCGGCAUUGUUCCUUUGUUUUUCGUCUUGGAUUUUUAAUACCCUUUGAUGGUAUCGUA-GG----CGCAAUGAGUGCU-AAGCACUUUGG--CUCGCUUAUUUAU (((((((..((((((.(((.((....(((((..((...........))...)))))...)).)-))----.))))))(((((.-..)))))...)--))))))....... ( -29.80, z-score = -2.05, R) >consensus AGUGGGCGGCAUUUUUCCUUUGUUUUUCGUCUUGGAUUUU_AAUACCCUUUGAUGGUAUCGGA_GG____CGCAAUGAGUGCU_AAGCACUUUGG__CUCGCUUAUUUAU ..((((((((............(((........)))......(((((.......)))))............))...(((((......))))).......))))))..... (-12.02 = -11.85 + -0.17)
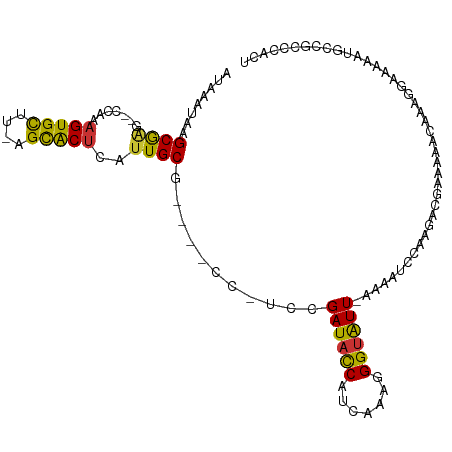
| Location | 12,766,057 – 12,766,159 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.11 |
| Shannon entropy | 0.40460 |
| G+C content | 0.43197 |
| Mean single sequence MFE | -22.10 |
| Consensus MFE | -11.44 |
| Energy contribution | -11.15 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.683147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
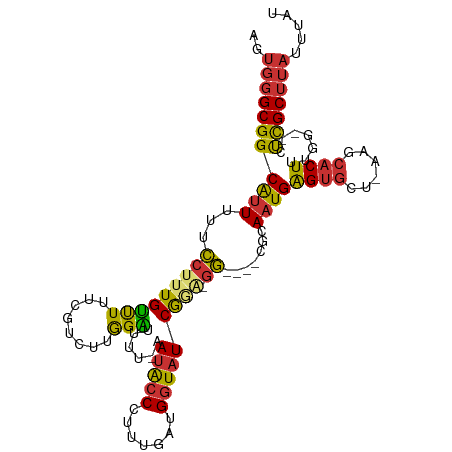
>dm3.chrX 12766057 102 - 22422827 AUAAAUAAGCGAG--CCAAAGUGCUU-AGCACUCAUUGCG----CCUUUCGAUACCAUCAAAGGGUAUU-AAAAUCCAAGACGAAAAACAAAGGAAAAAUGCCGCCCACU ........(((.(--(...(((((..-.))))).(((...----(((((.((((((.......))))))-....((......)).....)))))...))))))))..... ( -18.10, z-score = -0.58, R) >droWil1.scaffold_180588 858445 98 - 1294757 AUAAAUUAGCAGA--UUUAAGUGUUUCAGUGCCCAAUGCGCGUACCCCUUGAUAUCAUUAAAGGGCGUU---UUCCCAAACC--GAAACAAAGGCAAAAUGCCGC----- .((((((....))--))))..((((((.((((.....))))..(((((((..........))))).)).---..........--))))))..(((.....)))..----- ( -22.60, z-score = -1.50, R) >dp4.chrXL_group3a 1009780 95 + 2690836 AUAAAUAAGCGGC--CCAAAGUGUUU-AGCACUCAUUGCG----UCGCCCGAUACCAUCAAAUGGUAUU-AAGCCCCA------AAGGCAAUGAAAAAAUGCCGCCCAA- ........(((((--.....(((...-..)))(((((((.----(.((..((((((((...))))))))-..))....------.).)))))))......)))))....- ( -28.40, z-score = -3.81, R) >droPer1.super_52 134598 96 - 549863 AUAAAUAAGCGGC--CCAAAGUGUUU-AGCACUCAUUGCG----UAGCCCGAUACCAUCAAAUGGUAUU-AAGCCCUA------AAGGCAAUGAAAAAAUGCCGCCCAAC ........(((((--.....(((...-..)))(((((((.----(.((..((((((((...))))))))-..))....------.).)))))))......)))))..... ( -28.40, z-score = -3.91, R) >droEre2.scaffold_4690 4661630 103 + 18748788 AUAAAUAAGCGAGUGCCAAAGUGCUU-AGCACUCAUUGCG----CC-UCCGAUACCAUCAAAGGGUAUU-AAAAUCCAAGACGAAAAACAAAGGCAGAACGCCGCCCACU ........(((((.((((((((((..-.)))))..))).)----))-)).((((((.......))))))-......................(((.....)))))..... ( -22.40, z-score = -1.45, R) >droYak2.chrX 7042260 101 - 21770863 AUAAAUAAGCGAG--CCAAAGUGCUU-AGCACUCAUUGCG----CC-UCCGAUACCAUCAAAGGGUAUU-AAAAUCCAAGACGAAAAACAAAGGAAGAAUGCCGCCCACU ........(((.(--(((((((((..-.)))))..))).)----).-(((((((((.......))))))-....((......))........))).......)))..... ( -17.90, z-score = -0.52, R) >droSec1.super_21 516141 102 - 1102487 AUAAAUAAGCGAG--ACAAAGUGCUU-AGCACUCAUUGCG----CC-UACGAUACCAUCAAAGGGUAUUAAAAAUCCAAGACGAAAAACAAAGGAACAAUGCCGCCCACU ........(((..--....(((((..-.)))))(((((..----((-(..((((((.......)))))).....((......)).......)))..))))).)))..... ( -19.40, z-score = -1.71, R) >droSim1.chrX 9734191 102 - 17042790 AUAAAUAAGCGAG--CCAAAGUGCUU-AGCACUCAUUGCG----CC-UACGAUACCAUCAAAGGGUAUUAAAAAUCCAAGACGAAAAACAAAGGAACAAUGCCGCCCACU ........(((.(--(...(((((..-.))))).((((..----((-(..((((((.......)))))).....((......)).......)))..)))))))))..... ( -19.60, z-score = -1.40, R) >consensus AUAAAUAAGCGAG__CCAAAGUGCUU_AGCACUCAUUGCG____CC_UCCGAUACCAUCAAAGGGUAUU_AAAAUCCAAGACGAAAAACAAAGGAAAAAUGCCGCCCACU ........(((........(((((....))))).................((((((.......)))))).................................)))..... (-11.44 = -11.15 + -0.29)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:38:25 2011