| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,765,856 – 12,766,008 |
| Length | 152 |
| Max. P | 0.990460 |

| Location | 12,765,856 – 12,766,008 |
|---|---|
| Length | 152 |
| Sequences | 6 |
| Columns | 159 |
| Reading direction | reverse |
| Mean pairwise identity | 77.20 |
| Shannon entropy | 0.41244 |
| G+C content | 0.39088 |
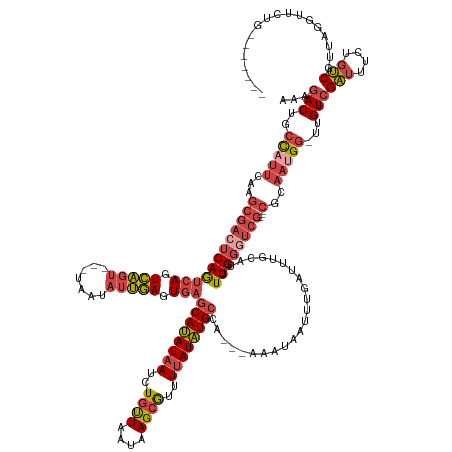
| Mean single sequence MFE | -40.42 |
| Consensus MFE | -24.01 |
| Energy contribution | -25.68 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.990460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
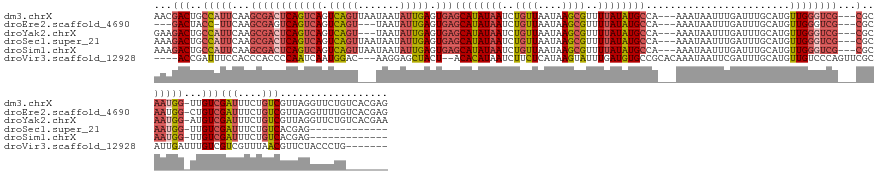
>dm3.chrX 12765856 152 - 22422827 AACGACUGCCAUUCAAGCGACUCAGUCAGUCAGUUAAUAAUAUUGAGUGAGCAUAUAAUCUGUUAAUAAGCGUUUUAUAUGCCA---AAAUAAUUUGAUUUGCAUGUUGGGUCG---CGCAAUGG-UUGUCGAUUUCUGUCGUUAGGUUCUGUCACGAG ..((((.((((((...(((((((((.((((.((((((...((((...((.((((((((..((((....))))..))))))))))---.))))..)))))).)).))))))))))---)..)))))-).)))).......(((((((...)))..)))). ( -47.60, z-score = -3.70, R) >droEre2.scaffold_4690 4661445 145 + 18748788 ---GACUACC-UUCAAGCGAGUCAGUCAGUCAGU---UAAUAUUGAGUGAGCAUAUAAUCUGUUAAUAAGCGUUUUAUAUGCCA---AAAUAAUUUGAUUUGCAUGUUGGGUCG---CGCAAUGG-CUGUCGAUUUCUGUCGUUAGGUUUUGUCACGAG ---.....((-..((.((((((((((((.(((((---....))))).)))((((((((..((((....))))..))))))))..---.......))))))))).))..)).(((---.((((.(.-(((.((((....)))).))).).))).).))). ( -42.70, z-score = -2.80, R) >droYak2.chrX 7042063 149 - 21770863 GAAGACUGCCAUUCAAGCGACUCAGUCAGUCAGU---UAAUAUUGAGUGAGCAUAUAAUCUGUUAAUAAGCGUUUUAUAUGCCA---AAAUAAUUUGAUUUGCAUGUUGGGUCG---CGCAAUGG-AUGUCGAUUUCUGUCGUUAGGUUCUGUCACGAA ...(((.((((((((.(((((((((.((((.(((---(((((((...((.((((((((..((((....))))..))))))))))---.))))..)))))).)).))))))))))---)....)))-))..((((....))))...)))...)))..... ( -43.50, z-score = -2.41, R) >droSec1.super_21 515982 139 - 1102487 AAAGACUGCCAUUCAAGCGACUCAGUCAGUCAGUUAAUAAUAUUGAGUGAGCAUAUAAUCUGUUAAUAAGCGUUUUAUAUGCCA---AAAUAAUUUGAUUUGCAUGUUGGGUCG---CGCAAUGG-UUGUCGAUUUCUGUCACGAG------------- ...(((.((((((...(((((((((.((((.((((((...((((...((.((((((((..((((....))))..))))))))))---.))))..)))))).)).))))))))))---)..)))))-).)))(((....))).....------------- ( -45.40, z-score = -4.47, R) >droSim1.chrX 9734032 139 - 17042790 AAAGACUGCCAUUCAAGCGACUCAGUCAGUCAGUUAAUAAUAUUGAGUGAGCAUAUAAUCUGUUAAUAAGCGUUUUAUAUGCCA---AAAUAAUUUGAUUUGCAUGUUGGGUCG---CGCAAUGG-UUGUCGAUUUCUGUCACGAG------------- ...(((.((((((...(((((((((.((((.((((((...((((...((.((((((((..((((....))))..))))))))))---.))))..)))))).)).))))))))))---)..)))))-).)))(((....))).....------------- ( -45.40, z-score = -4.47, R) >droVir3.scaffold_12928 3134756 143 + 7717345 ----ACCGAUUUCCACCCACCCCAAUCAAUGGAC---AAGGAGCUACU--ACACAUAAUCUUCUCAUAAGUAUUUGAUGUGCCGCACAAAUAAUUCGAUUUGCAUGUUGUCCCAGUUCGCAUUGAUUUGUCGUCGUUUAACGUUCUACCCUG------- ----..((((.............((((((((((.---..((..(.((.--.(((((......((....))......))))).....(((((......)))))...)).)..))...)).)))))))).))))....................------- ( -17.94, z-score = 1.93, R) >consensus AAAGACUGCCAUUCAAGCGACUCAGUCAGUCAGU___UAAUAUUGAGUGAGCAUAUAAUCUGUUAAUAAGCGUUUUAUAUGCCA___AAAUAAUUUGAUUUGCAUGUUGGGUCG___CGCAAUGG_UUGUCGAUUUCUGUCGUUAGGUUCUG_______ ...(((..(((((....(((((((((((.(((((.......))))).)))((((((((..((((....))))..))))))))........................))))))))......)))))...)))(((....))).................. (-24.01 = -25.68 + 1.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:38:23 2011