| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,229,720 – 11,229,823 |
| Length | 103 |
| Max. P | 0.808495 |

| Location | 11,229,720 – 11,229,823 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.24 |
| Shannon entropy | 0.57172 |
| G+C content | 0.36646 |
| Mean single sequence MFE | -21.65 |
| Consensus MFE | -9.27 |
| Energy contribution | -9.54 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.808495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
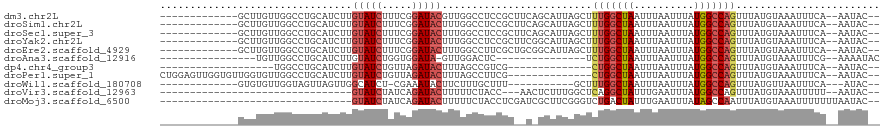
>dm3.chr2L 11229720 103 + 23011544 -------------GCUUGUUGGCCUGCAUCUUGUAUCUUUCGGAUACGUUGGCCUCCGCUUCAGCAUUAGCUUUGGCUAAUUUAAUUUAUGGCCAGUUUAUGUAAAUUUCA--AAUAC-- -------------.....((((((........(((((.....)))))(((((((...(((........)))...))))))).........))))))...............--.....-- ( -25.90, z-score = -1.78, R) >droSim1.chr2L 11029105 103 + 22036055 -------------GCUUGUUGGCCUGCAUCUUGUAUCUUUCGGAUACUUUGGCCUCCGCUUCAGCAUUAGCUUUGGCUAAUUUAAUUUAUGGCCAGUUUAUGUAAAUUUCA--AAUAC-- -------------.....((((((........(((((.....))))).((((((...(((........)))...))))))..........))))))...............--.....-- ( -24.40, z-score = -1.51, R) >droSec1.super_3 6626547 103 + 7220098 -------------GCUUGUUGGCCUGCAUCUUGUAUCUUUCGGAUACUUUGGCCUCCGCUUCAGCAUUAGCUUUGGCUAAUUUAAUUUAUGGCCAGUUUAUGUAAAUUUCA--AAUAC-- -------------.....((((((........(((((.....))))).((((((...(((........)))...))))))..........))))))...............--.....-- ( -24.40, z-score = -1.51, R) >droYak2.chr2L 7625541 103 + 22324452 -------------GCUUGUUGGCCUGCAUCUUGUAUCUUUCGGAUACUUUGGCCUCCGCUUCGGCAUUAGCUUUGGCUAAUUUAAUUUAUGGCCAGUUUAUGUAAAUUUCA--AAUAC-- -------------(((....))).(((((...(((((.....))))).((((((...((....))((((((....)))))).........))))))...))))).......--.....-- ( -27.30, z-score = -2.35, R) >droEre2.scaffold_4929 12424267 103 - 26641161 -------------GCUUGUUGGCCUGCAUCUUGUAUCUUUCGGAUACUUUGGCCUUCGCUGCGGCAUUAGCUUUGGCUAAUUUAAUUUAUGGCCAGUUUAUGUAAAUUUCA--AAUAC-- -------------.....((((((........(((((.....))))).((((((...((((......))))...))))))..........))))))...............--.....-- ( -25.00, z-score = -1.14, R) >droAna3.scaffold_12916 11952001 86 - 16180835 ----------------UGUUGGCCUGCAUCUUGUAUCUGGUGGAUA-GUUGGACUC---------------UCUGGCUAAUUUAAUUUAUGGCCAGUUUAUGUAAAUUUCG--AAAAUAC ----------------..((((((..((((........))))..((-((..(....---------------.)..))))...........))))))...............--....... ( -19.00, z-score = -1.12, R) >dp4.chr4_group3 6644570 83 + 11692001 -------------------UGGCCUGCAUCUUGUAUCUGUUAGAUACUUUAGCCGUCG--------------CUGGCUAAUUUAAUUUAUGGCCAGUUUAUGUAAAUUUCA--AAUAC-- -------------------(((((........(((((.....))))).(((((((...--------------.)))))))..........)))))................--.....-- ( -20.70, z-score = -2.29, R) >droPer1.super_1 8099817 102 + 10282868 CUGGAGUUGGUGUUGGUGUUGGCCUGCAUCUUGUAUCUGUUAGAUACUUUAGCCUUCG--------------CUGGCUAAUUUAAUUUAUGGCCAGUUUAUGUAAAUUUCA--AAUAC-- .((((((((((((.(((....))).)))))..(((((.....)))))....((.(..(--------------(((((((..........))))))))..).)).)))))))--.....-- ( -26.40, z-score = -1.73, R) >droWil1.scaffold_180708 8026908 90 - 12563649 -------------GUGUGUUGGUAGUUAGUUGGCAUCU-CGAAAUACUUCUUUGCUUU-----------GCUUUGGCUAAUUUAAUUUAUGGCCAGUUUAUGUUAAUUUCA---AUAC-- -------------..(((((((.((((((..((((...-((((.......))))...)-----------)))(((((((..........)))))))......)))))))))---))))-- ( -17.30, z-score = -0.92, R) >droVir3.scaffold_12963 17928538 81 - 20206255 --------------------------------GUAUCUAUCAGAUACUUUUUCUACC---AACUCUUUGGCUCAGGCUAUUUGAAUUUAUGGCCAGUUUAUGUAAAUUUUU--AAUAC-- --------------------------------(((((.....)))))........((---((....))))....((((((........)))))).................--.....-- ( -14.10, z-score = -1.67, R) >droMoj3.scaffold_6500 4855811 86 - 32352404 --------------------------------GUAUCUAUCAGAUACUUUUUCUACCUCGAUCGCUUCGGGUCUGACUAUUUGAAUUUAUAGCCAAUUUAUGUAAAUUUUUUUAAUAC-- --------------------------------(((((.....)))))....((.(((.(((.....))))))..)).((((.(((((((((.........)))))))))....)))).-- ( -13.70, z-score = -1.67, R) >consensus _____________GCUUGUUGGCCUGCAUCUUGUAUCUUUCGGAUACUUUGGCCUCCGC__C_GCAUUAGCUUUGGCUAAUUUAAUUUAUGGCCAGUUUAUGUAAAUUUCA__AAUAC__ ................................(((((.....))))).........................(((((((..........)))))))........................ ( -9.27 = -9.54 + 0.27)

| Location | 11,229,720 – 11,229,823 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.24 |
| Shannon entropy | 0.57172 |
| G+C content | 0.36646 |
| Mean single sequence MFE | -19.33 |
| Consensus MFE | -6.58 |
| Energy contribution | -6.51 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.772426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
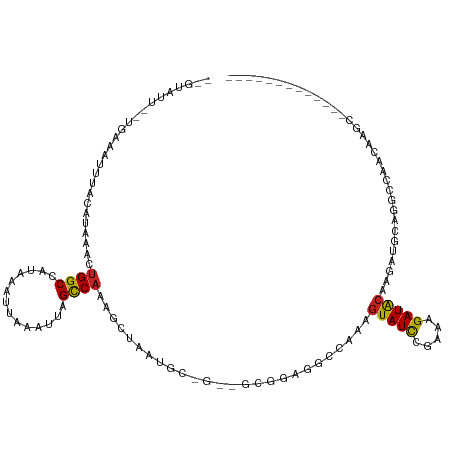
>dm3.chr2L 11229720 103 - 23011544 --GUAUU--UGAAAUUUACAUAAACUGGCCAUAAAUUAAAUUAGCCAAAGCUAAUGCUGAAGCGGAGGCCAACGUAUCCGAAAGAUACAAGAUGCAGGCCAACAAGC------------- --(((((--(...............(((((.........((((((....))))))((....))...)))))..((((.(....))))).))))))............------------- ( -24.60, z-score = -2.24, R) >droSim1.chr2L 11029105 103 - 22036055 --GUAUU--UGAAAUUUACAUAAACUGGCCAUAAAUUAAAUUAGCCAAAGCUAAUGCUGAAGCGGAGGCCAAAGUAUCCGAAAGAUACAAGAUGCAGGCCAACAAGC------------- --(((((--(...............(((((.........((((((....))))))((....))...)))))..((((.(....))))).))))))............------------- ( -24.50, z-score = -2.25, R) >droSec1.super_3 6626547 103 - 7220098 --GUAUU--UGAAAUUUACAUAAACUGGCCAUAAAUUAAAUUAGCCAAAGCUAAUGCUGAAGCGGAGGCCAAAGUAUCCGAAAGAUACAAGAUGCAGGCCAACAAGC------------- --(((((--(...............(((((.........((((((....))))))((....))...)))))..((((.(....))))).))))))............------------- ( -24.50, z-score = -2.25, R) >droYak2.chr2L 7625541 103 - 22324452 --GUAUU--UGAAAUUUACAUAAACUGGCCAUAAAUUAAAUUAGCCAAAGCUAAUGCCGAAGCGGAGGCCAAAGUAUCCGAAAGAUACAAGAUGCAGGCCAACAAGC------------- --(((((--(...............(((((.........((((((....)))))).(((...))).)))))..((((.(....))))).))))))............------------- ( -24.60, z-score = -2.47, R) >droEre2.scaffold_4929 12424267 103 + 26641161 --GUAUU--UGAAAUUUACAUAAACUGGCCAUAAAUUAAAUUAGCCAAAGCUAAUGCCGCAGCGAAGGCCAAAGUAUCCGAAAGAUACAAGAUGCAGGCCAACAAGC------------- --.....--................(((((.........((((((....))))))...(((((....))....((((.(....)))))....))).)))))......------------- ( -25.60, z-score = -2.93, R) >droAna3.scaffold_12916 11952001 86 + 16180835 GUAUUUU--CGAAAUUUACAUAAACUGGCCAUAAAUUAAAUUAGCCAGA---------------GAGUCCAAC-UAUCCACCAGAUACAAGAUGCAGGCCAACA---------------- (((((((--.(.............(((((..............))))).---------------.........-((((.....)))))))))))).........---------------- ( -11.34, z-score = -0.79, R) >dp4.chr4_group3 6644570 83 - 11692001 --GUAUU--UGAAAUUUACAUAAACUGGCCAUAAAUUAAAUUAGCCAG--------------CGACGGCUAAAGUAUCUAACAGAUACAAGAUGCAGGCCA------------------- --.....--.................((((..........((((((..--------------....)))))).(((((.....)))))........)))).------------------- ( -21.70, z-score = -3.60, R) >droPer1.super_1 8099817 102 - 10282868 --GUAUU--UGAAAUUUACAUAAACUGGCCAUAAAUUAAAUUAGCCAG--------------CGAAGGCUAAAGUAUCUAACAGAUACAAGAUGCAGGCCAACACCAACACCAACUCCAG --.....--................(((((..........((((((..--------------....)))))).(((((.....)))))........)))))................... ( -22.50, z-score = -3.62, R) >droWil1.scaffold_180708 8026908 90 + 12563649 --GUAU---UGAAAUUAACAUAAACUGGCCAUAAAUUAAAUUAGCCAAAGC-----------AAAGCAAAGAAGUAUUUCG-AGAUGCCAACUAACUACCAACACAC------------- --((((---((((((..((......((((..............))))..((-----------...))......))))))).-.)))))...................------------- ( -7.34, z-score = 0.42, R) >droVir3.scaffold_12963 17928538 81 + 20206255 --GUAUU--AAAAAUUUACAUAAACUGGCCAUAAAUUCAAAUAGCCUGAGCCAAAGAGUU---GGUAGAAAAAGUAUCUGAUAGAUAC-------------------------------- --.....--.....(((((...(((((((((...............)).)))....))))---.)))))....(((((.....)))))-------------------------------- ( -10.86, z-score = -0.57, R) >droMoj3.scaffold_6500 4855811 86 + 32352404 --GUAUUAAAAAAAUUUACAUAAAUUGGCUAUAAAUUCAAAUAGUCAGACCCGAAGCGAUCGAGGUAGAAAAAGUAUCUGAUAGAUAC-------------------------------- --......................((((((((........))))))))((((((.....))).))).......(((((.....)))))-------------------------------- ( -15.10, z-score = -2.24, R) >consensus __GUAUU__UGAAAUUUACAUAAACUGGCCAUAAAUUAAAUUAGCCAAAGCUAAUGC_G__GCGGAGGCCAAAGUAUCCGAAAGAUACAAGAUGCAGGCCAACAAGC_____________ .........................((((..............))))..........................(((((.....)))))................................ ( -6.58 = -6.51 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:53 2011