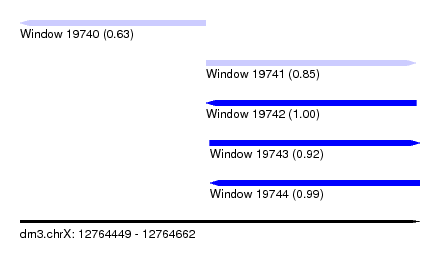
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,764,449 – 12,764,662 |
| Length | 213 |
| Max. P | 0.997461 |

| Location | 12,764,449 – 12,764,548 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Shannon entropy | 0.02755 |
| G+C content | 0.42135 |
| Mean single sequence MFE | -24.57 |
| Consensus MFE | -22.29 |
| Energy contribution | -22.73 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.627982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12764449 99 - 22422827 GCAAUGCACUUUUUCCGGCCAUCUGGUU-GGAAUACUGGUCCAUAAAGUUUAAUGAUCCAUACACAACGAAGCUCCUAAAGUUAGAUUUCGGCAAAACUU ....(((.....((((((((....))))-))))...(((..(((........)))..))).......(((((...(((....))).))))))))...... ( -19.90, z-score = -0.59, R) >droSec1.super_21 514680 100 - 1102487 GCAAUGCACUUUUUCCGGCCAUCUGGCUGGGAAUACUGGUCCAUAAAGUUUAAUGAUCCAUACACAACGAAGCUCCUAAAGUGAGAUUUCGGCAAAACUU ....(((....(((((((((....)))))))))...(((..(((........)))..))).......(((((((((....).))).))))))))...... ( -26.90, z-score = -2.12, R) >droSim1.chrX 9732726 100 - 17042790 GCAAUGCACUUUUUCCGGCCAUCUGGCUGGGAAUACUGGUCCAUAAAGUUUAAUGAUCCAUACACAACGAAGCUCCUAAAGUGAGAUUUCGGCAAAACUU ....(((....(((((((((....)))))))))...(((..(((........)))..))).......(((((((((....).))).))))))))...... ( -26.90, z-score = -2.12, R) >consensus GCAAUGCACUUUUUCCGGCCAUCUGGCUGGGAAUACUGGUCCAUAAAGUUUAAUGAUCCAUACACAACGAAGCUCCUAAAGUGAGAUUUCGGCAAAACUU ....(((....(((((((((....)))))))))...(((..(((........)))..))).......((((((((.......))).))))))))...... (-22.29 = -22.73 + 0.45)

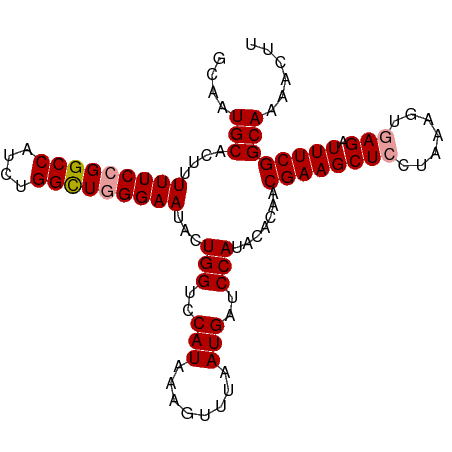
| Location | 12,764,548 – 12,764,660 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 67.66 |
| Shannon entropy | 0.56796 |
| G+C content | 0.35859 |
| Mean single sequence MFE | -23.14 |
| Consensus MFE | -8.24 |
| Energy contribution | -8.76 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.848168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12764548 112 + 22422827 UGCAUGGA-----CUUUCCCAAACAAAAUCAUAUCAGAUGUCACGAUGUAAUUUGGAGUAUUUCUAUAGAAUAUCCUUUCUGACAUCCUAGACUUAAAAA-UACCGCUCUACCGAAAA .((.(((.-----.....)))..................(((..(((((.....(((....(((....)))..)))......)))))...))).......-....))........... ( -15.90, z-score = -0.22, R) >droEre2.scaffold_4690 4660200 102 - 18748788 UGCAUGAU-----AUA--------GACAGGAUAUCAAAUGUCAUGAUGUCAUUUGGAGUAUUCCUAUAGAAUGUCCUUUUAGACAUUCUAGACUAGAAUAGCCCCCCUUUACUGA--- ........-----...--------..((((((((((.......)))))))....((..(((((...(((((((((......))))))))).....)))))..)).......))).--- ( -25.40, z-score = -2.28, R) >droSec1.super_21 514780 110 + 1102487 UGCAUGGA-----CUUUCCCAAACAACAUCAUAUCAGAUGUCAUGAUGCCAUUUGGAGUAUUUCUAUAGAAUGUCCUUUCAGACAUUCUAGACUAGAAUA---CCGCUCUACCGAAAA .((((((.-----.....)).....(((((......)))))....))))....((((((..((((((((((((((......)))))))))...)))))..---..))))))....... ( -27.50, z-score = -3.04, R) >droSim1.chrX 9732826 110 + 17042790 UGCACGGA-----CUUUCCCAAACAACAUCAUAUCAGAUGUCAUGAUGUCAUUUCGAGUAUUUCUAUAGAAUGUCCUUUCAGACAUUCUAGAAUAGAAUA---CCGCUCUACCGAAAA ....(((.-----............(((((((..(....)..)))))))......((((..((((((((((((((......)))))))))...)))))..---..))))..))).... ( -27.70, z-score = -3.63, R) >triCas2.chrUn_29 118726 113 + 145421 UACGUGAUUAUCGGUCCACCAUUUGAAAAAAUAACAGUUGGCUUCAUUAAGGCCCUAGACUUUCUAAUUAACCUUGUUUUUUACAAAAAUCAAUAAAAU-----CGCUACACGGAAAA ...((((((...((....)).(((((((((((((..((((((((.....))))).(((.....)))...))).))))))))).)))).........)))-----)))........... ( -19.20, z-score = -1.26, R) >consensus UGCAUGGA_____CUUUCCCAAACAACAUCAUAUCAGAUGUCAUGAUGUCAUUUGGAGUAUUUCUAUAGAAUGUCCUUUCAGACAUUCUAGACUAGAAUA___CCGCUCUACCGAAAA .................................(((((((.(.....).)))))))..........(((((((((......)))))))))............................ ( -8.24 = -8.76 + 0.52)


| Location | 12,764,548 – 12,764,660 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 67.66 |
| Shannon entropy | 0.56796 |
| G+C content | 0.35859 |
| Mean single sequence MFE | -30.14 |
| Consensus MFE | -15.96 |
| Energy contribution | -17.52 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.997461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12764548 112 - 22422827 UUUUCGGUAGAGCGGUA-UUUUUAAGUCUAGGAUGUCAGAAAGGAUAUUCUAUAGAAAUACUCCAAAUUACAUCGUGACAUCUGAUAUGAUUUUGUUUGGGAAAG-----UCCAUGCA ......((((..(((((-((((((....(((((((((......)))))))))))))))))))((((((...((((((.(....).))))))...))))))....)-----..).))). ( -27.60, z-score = -1.70, R) >droEre2.scaffold_4690 4660200 102 + 18748788 ---UCAGUAAAGGGGGGCUAUUCUAGUCUAGAAUGUCUAAAAGGACAUUCUAUAGGAAUACUCCAAAUGACAUCAUGACAUUUGAUAUCCUGUC--------UAU-----AUCAUGCA ---...((.....((((.((((((....((((((((((....))))))))))..)))))))))).....))..(((((.((..(((.....)))--------.))-----.))))).. ( -27.60, z-score = -2.29, R) >droSec1.super_21 514780 110 - 1102487 UUUUCGGUAGAGCGG---UAUUCUAGUCUAGAAUGUCUGAAAGGACAUUCUAUAGAAAUACUCCAAAUGGCAUCAUGACAUCUGAUAUGAUGUUGUUUGGGAAAG-----UCCAUGCA ......((((..(((---(((((((...((((((((((....))))))))))))).))))))(((((..((((((((.(....).))))))))..)))))....)-----..).))). ( -40.40, z-score = -4.98, R) >droSim1.chrX 9732826 110 - 17042790 UUUUCGGUAGAGCGG---UAUUCUAUUCUAGAAUGUCUGAAAGGACAUUCUAUAGAAAUACUCGAAAUGACAUCAUGACAUCUGAUAUGAUGUUGUUUGGGAAAG-----UCCGUGCA .........(.((((---..(((((...((((((((((....)))))))))))))))...(((.(((..((((((((.(....).))))))))..))))))....-----.)))).). ( -36.90, z-score = -4.12, R) >triCas2.chrUn_29 118726 113 - 145421 UUUUCCGUGUAGCG-----AUUUUAUUGAUUUUUGUAAAAAACAAGGUUAAUUAGAAAGUCUAGGGCCUUAAUGAAGCCAACUGUUAUUUUUUCAAAUGGUGGACCGAUAAUCACGUA .....((((...((-----.(((((((((...((((.....))))(((...((((.....)))).))))))))))))....(..((((((....))))))..)..)).....)))).. ( -18.20, z-score = 0.75, R) >consensus UUUUCGGUAGAGCGG___UAUUCUAGUCUAGAAUGUCUGAAAGGACAUUCUAUAGAAAUACUCCAAAUGACAUCAUGACAUCUGAUAUGAUGUUGUUUGGGAAAG_____UCCAUGCA ...........(((......(((((...((((((((((....))))))))))))))).......(((((((((((((.(....).)))))))))))))................))). (-15.96 = -17.52 + 1.56)

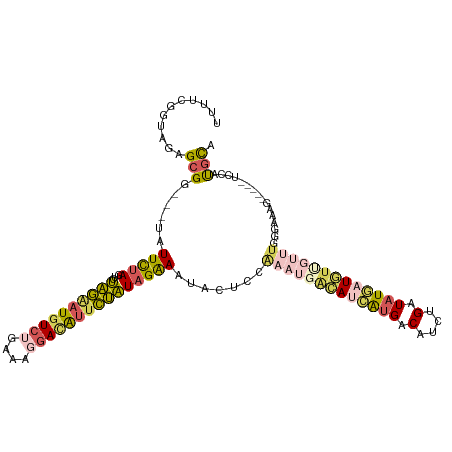
| Location | 12,764,550 – 12,764,662 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 65.76 |
| Shannon entropy | 0.59963 |
| G+C content | 0.35981 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -8.24 |
| Energy contribution | -8.76 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12764550 112 + 22422827 -----CAUGGACUUUCCCAAACA-AAAUCAUAUCAGAUGUCACGAUGUAAUUUGGAGUAUUUCUAUAGAAUAUCCUUUCUGACAUCCUAGACUUAAAAA-UACCGCUCUACCGAAAAAC -----..(((.............-..(((......)))(((..(((((.....(((....(((....)))..)))......)))))...))).......-..........)))...... ( -14.20, z-score = 0.15, R) >droEre2.scaffold_4690 4660202 100 - 18748788 --------------CAUGAUAUAGACAGGAUAUCAAAUGUCAUGAUGUCAUUUGGAGUAUUCCUAUAGAAUGUCCUUUUAGACAUUCUAGACUAGAAUAGCCCCCCUUUACUGA----- --------------...........((((((((((.......)))))))....((..(((((...(((((((((......))))))))).....)))))..)).......))).----- ( -25.40, z-score = -2.47, R) >droSec1.super_21 514782 110 + 1102487 -----CAUGGACUUUCCCAAACA-ACAUCAUAUCAGAUGUCAUGAUGCCAUUUGGAGUAUUUCUAUAGAAUGUCCUUUCAGACAUUCUAGACUAGAAUA---CCGCUCUACCGAAAAAC -----..(((......(((((..-.((((((..(....)..))))))...))))).(((((.((((((((((((......)))))))))...)))))))---).......)))...... ( -26.50, z-score = -3.10, R) >droSim1.chrX 9732828 110 + 17042790 -----CACGGACUUUCCCAAACA-ACAUCAUAUCAGAUGUCAUGAUGUCAUUUCGAGUAUUUCUAUAGAAUGUCCUUUCAGACAUUCUAGAAUAGAAUA---CCGCUCUACCGAAAAAC -----..(((.............-(((((((..(....)..)))))))......((((..((((((((((((((......)))))))))...)))))..---..))))..)))...... ( -27.70, z-score = -3.99, R) >triCas2.chrUn_29 118728 113 + 145421 CGUGAUUAUCGGUCCACCAUUUG-AAAAAAUAACAGUUGGCUUCAUUAAGGCCCUAGACUUUCUAAUUAACCUUGUUUUUUACAAAAAUCAAUAAAAU-----CGCUACACGGAAAAAC .((((((...((....)).((((-(((((((((..((((((((.....))))).(((.....)))...))).))))))))).)))).........)))-----)))............. ( -19.20, z-score = -1.28, R) >consensus _____CAUGGACUUUCCCAAACA_ACAUCAUAUCAGAUGUCAUGAUGUCAUUUGGAGUAUUUCUAUAGAAUGUCCUUUCAGACAUUCUAGACUAGAAUA___CCGCUCUACCGAAAAAC ................................(((((((.(.....).)))))))..........(((((((((......))))))))).............................. ( -8.24 = -8.76 + 0.52)


| Location | 12,764,550 – 12,764,662 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 65.76 |
| Shannon entropy | 0.59963 |
| G+C content | 0.35981 |
| Mean single sequence MFE | -29.64 |
| Consensus MFE | -12.82 |
| Energy contribution | -12.90 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.992984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12764550 112 - 22422827 GUUUUUCGGUAGAGCGGUA-UUUUUAAGUCUAGGAUGUCAGAAAGGAUAUUCUAUAGAAAUACUCCAAAUUACAUCGUGACAUCUGAUAUGAUUU-UGUUUGGGAAAGUCCAUG----- ..(((((........((((-((((((....(((((((((......)))))))))))))))))))((((((...((((((.(....).))))))..-.)))))))))))......----- ( -27.00, z-score = -1.60, R) >droEre2.scaffold_4690 4660202 100 + 18748788 -----UCAGUAAAGGGGGGCUAUUCUAGUCUAGAAUGUCUAAAAGGACAUUCUAUAGGAAUACUCCAAAUGACAUCAUGACAUUUGAUAUCCUGUCUAUAUCAUG-------------- -----...((.....((((.((((((....((((((((((....))))))))))..)))))))))).....))..(((((.((..(((.....))).)).)))))-------------- ( -27.20, z-score = -2.39, R) >droSec1.super_21 514782 110 - 1102487 GUUUUUCGGUAGAGCGG---UAUUCUAGUCUAGAAUGUCUGAAAGGACAUUCUAUAGAAAUACUCCAAAUGGCAUCAUGACAUCUGAUAUGAUGU-UGUUUGGGAAAGUCCAUG----- ..(((((........((---(((((((...((((((((((....))))))))))))).))))))(((((..((((((((.(....).))))))))-..))))))))))......----- ( -39.80, z-score = -5.00, R) >droSim1.chrX 9732828 110 - 17042790 GUUUUUCGGUAGAGCGG---UAUUCUAUUCUAGAAUGUCUGAAAGGACAUUCUAUAGAAAUACUCGAAAUGACAUCAUGACAUCUGAUAUGAUGU-UGUUUGGGAAAGUCCGUG----- .............((((---..(((((...((((((((((....)))))))))))))))...(((.(((..((((((((.(....).))))))))-..)))))).....)))).----- ( -36.60, z-score = -4.15, R) >triCas2.chrUn_29 118728 113 - 145421 GUUUUUCCGUGUAGCG-----AUUUUAUUGAUUUUUGUAAAAAACAAGGUUAAUUAGAAAGUCUAGGGCCUUAAUGAAGCCAACUGUUAUUUUUU-CAAAUGGUGGACCGAUAAUCACG .......((((...((-----.(((((((((...((((.....))))(((...((((.....)))).))))))))))))....(..((((((...-.))))))..)..)).....)))) ( -17.60, z-score = 0.90, R) >consensus GUUUUUCGGUAGAGCGG___UAUUCUAGUCUAGAAUGUCUGAAAGGACAUUCUAUAGAAAUACUCCAAAUGACAUCAUGACAUCUGAUAUGAUGU_UGUUUGGGAAAGUCCAUG_____ ......................(((((...((((((((((....)))))))))))))))............((((((((.(....).))))))))........................ (-12.82 = -12.90 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:38:22 2011