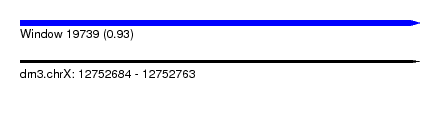
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,752,684 – 12,752,763 |
| Length | 79 |
| Max. P | 0.927195 |

| Location | 12,752,684 – 12,752,763 |
|---|---|
| Length | 79 |
| Sequences | 4 |
| Columns | 80 |
| Reading direction | forward |
| Mean pairwise identity | 74.34 |
| Shannon entropy | 0.38891 |
| G+C content | 0.41181 |
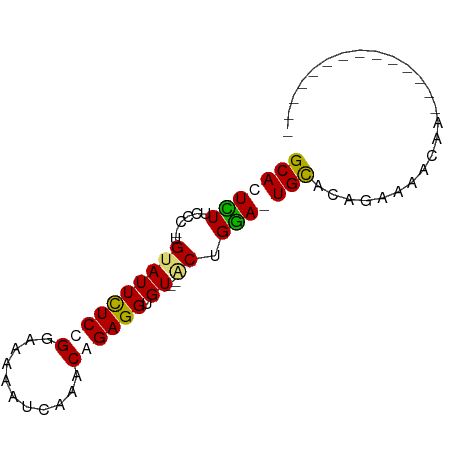
| Mean single sequence MFE | -16.10 |
| Consensus MFE | -10.78 |
| Energy contribution | -10.72 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.927195 |
| Prediction | RNA |
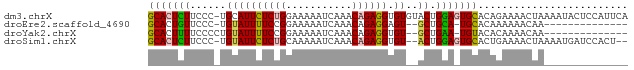
Download alignment: ClustalW | MAF
>dm3.chrX 12752684 79 + 22422827 GCACUCUUCCC-UGCAUUCUCUGGAAAAAUCAAACAGAGGUGUGUACUGGAGUGCACAGAAAACUAAAAUACUCCAUUCA (((((((....-(((((((((((...........)))))).)))))..)))))))......................... ( -22.00, z-score = -3.10, R) >droEre2.scaffold_4690 4648343 62 - 18748788 GCACUGUUCCC-UGUAUUUUCCGGAAAAAUCAAACAGAGGAGU--GCUGCA-UGCACAAAAAACAA-------------- .......((((-((((((((.....)))))...)))).)))((--((....-.)))).........-------------- ( -12.30, z-score = -0.72, R) >droYak2.chrX 7029173 63 + 21770863 GCACUUUUCCCCUGUAUUUUCCGGAAAAAUCAAACAGAGGUGU--GCUGAA-UGUACACAAAACAA-------------- ....((((((............))))))...........((((--((....-.)))))).......-------------- ( -11.10, z-score = -0.60, R) >droSim1.chrX 9720823 75 + 17042790 GCACUCUUCCC-UGUAUUCUCUGCAAAAAUCAAACAGAGGUGU--ACUGGAGUGCACUGAAAACUAAAAUGAUCCACU-- (((((((....-.((((((((((...........)))))).))--)).))))))).......................-- ( -19.00, z-score = -3.04, R) >consensus GCACUCUUCCC_UGUAUUCUCCGGAAAAAUCAAACAGAGGUGU__ACUGGA_UGCACAGAAAACAA______________ (((((((..........((((((...........))))))........)))))))......................... (-10.78 = -10.72 + -0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:38:18 2011