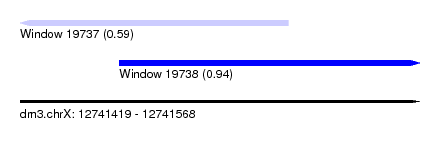
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,741,419 – 12,741,568 |
| Length | 149 |
| Max. P | 0.935759 |

| Location | 12,741,419 – 12,741,519 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 96.58 |
| Shannon entropy | 0.06024 |
| G+C content | 0.46776 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -26.86 |
| Energy contribution | -26.54 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
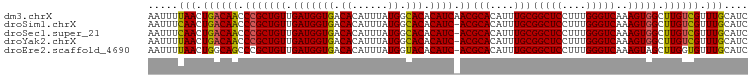
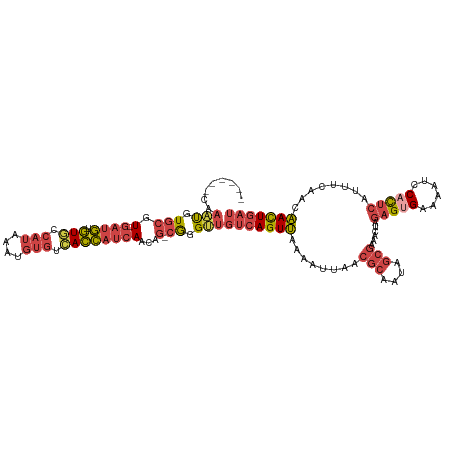
>dm3.chrX 12741419 100 - 22422827 AAUUUUAACUGACAACCCGCUGUUGAUGGUGACACAUUUAUGGCACACAUCAACGCACAUUUGCGGCUCCUUUGGGUCAAAGUGGCUUGUCGUUUGCAUC .....(((.((((((.(((((((((((((((.((......)).))).)))))))(((....)))(((((....)))))..))))).)))))).))).... ( -32.70, z-score = -2.75, R) >droSim1.chrX 9706157 99 - 17042790 AAUUUCAACUGACAACCCGCUGUUGAUGGUGACACAUUUAUGGCACACAUC-ACGCACAUUUGCGGCUCCUUUGGGUCAAAGUGGCUUGUCGUUUGCAUC .....(((.((((((.(((((..((((((((.((......)).))).))))-).(((....)))(((((....)))))..))))).)))))).))).... ( -29.50, z-score = -1.53, R) >droSec1.super_21 492268 99 - 1102487 AAUUUCAACUGACAACCCGCUGUUGAUGGUGACACAUUUAUGGCACACAUC-ACGCACAUUUGCGGCUCCUUUGGGUCAAAGUGGCUUGUCGUUUGCAUC .....(((.((((((.(((((..((((((((.((......)).))).))))-).(((....)))(((((....)))))..))))).)))))).))).... ( -29.50, z-score = -1.53, R) >droYak2.chrX 7017864 99 - 21770863 AAUUUUAACUGACAACCCGCUGUUGAUGGUGACACAUUUAUGGCACACAUC-ACGCACAUUUGCGGCUCCUUUGGGUCAAAGUGGCUUGUCGUUUGCAUC .....(((.((((((.(((((..((((((((.((......)).))).))))-).(((....)))(((((....)))))..))))).)))))).))).... ( -28.10, z-score = -1.29, R) >droEre2.scaffold_4690 4637393 99 + 18748788 AAUUUUAACUGGCAGCCCGCUGUUGAUGGUGACACAUUUAUGGUACACAUC-ACGCACAUUUGCGGCUCCUUUGGGUCAAAGUAGCUUGGUGUUUGCAUC ....(((((.(((.....))))))))((....))........(((.(((((-(((((....))))(((.(((((...))))).))).)))))).)))... ( -25.50, z-score = 0.15, R) >consensus AAUUUUAACUGACAACCCGCUGUUGAUGGUGACACAUUUAUGGCACACAUC_ACGCACAUUUGCGGCUCCUUUGGGUCAAAGUGGCUUGUCGUUUGCAUC .....(((.((((((.(((((((.(((((((.((......)).))).)))).))(((....)))(((((....)))))..))))).)))))).))).... (-26.86 = -26.54 + -0.32)
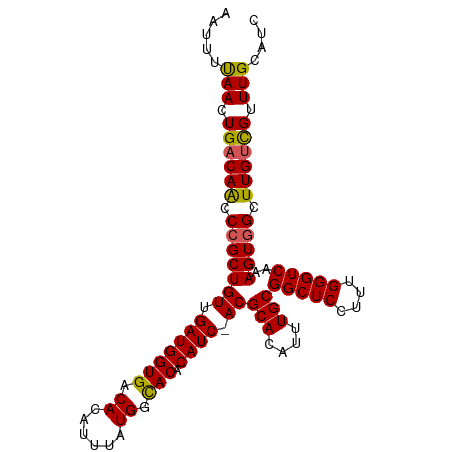
| Location | 12,741,456 – 12,741,568 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.82 |
| Shannon entropy | 0.39617 |
| G+C content | 0.38572 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -15.03 |
| Energy contribution | -16.87 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12741456 112 + 22422827 ------CAAAUGUGCGUUGAUGUGUGCCAUAAAUGUGUCACCAUCAACAG-CGGGUUGUCAGUUAAAAUUAACGCAAUAGCGAAACUAGAGUGCAAAUCCACUCAUUUCAACAACUGAU ------..(((.((((((((((.(((.(((....))).)))))))))).)-)).)))(((((((........(((....)))......(((((......)))))........))))))) ( -35.20, z-score = -3.70, R) >droSim1.chrX 9706194 111 + 17042790 ------CAAAUGUGCGU-GAUGUGUGCCAUAAAUGUGUCACCAUCAACAG-CGGGUUGUCAGUUGAAAUUAACGCAAUAGCGAAACUAGAGUGAAAAUCCACUCAUUUCAACAACUGAU ------..(((.(((.(-((((.(((.(((....))).))))))))...)-)).)))((((((((.......(((....)))......(((((......))))).......)))))))) ( -36.30, z-score = -3.94, R) >droSec1.super_21 492305 111 + 1102487 ------CAAAUGUGCGU-GAUGUGUGCCAUAAAUGUGUCACCAUCAACAG-CGGGUUGUCAGUUGAAAUUAACGCAAUAGCGAAACUAGAGUGAAAAUCCACUCAUUUCAACAACUGAU ------..(((.(((.(-((((.(((.(((....))).))))))))...)-)).)))((((((((.......(((....)))......(((((......))))).......)))))))) ( -36.30, z-score = -3.94, R) >droYak2.chrX 7017901 111 + 21770863 ------CAAAUGUGCGU-GAUGUGUGCCAUAAAUGUGUCACCAUCAACAG-CGGGUUGUCAGUUAAAAUUAACGCAAUAGCGAAACUAGAGUGAAAAUCCACACAUUUCAACAACUGAU ------..(((.(((.(-((((.(((.(((....))).))))))))...)-)).)))(((((((........(((....)))........(((......)))..........))))))) ( -28.60, z-score = -1.80, R) >droEre2.scaffold_4690 4637430 111 - 18748788 ------CAAAUGUGCGU-GAUGUGUACCAUAAAUGUGUCACCAUCAACAG-CGGGCUGCCAGUUAAAAUUAACGCAAUAGCGAAACUAGAGGGAAAAUCCACUCAUUUCAGAAACUGAU ------....(.(((.(-((((.((..(((....)))..)))))))...)-)).)....(((((........(((....)))......(((((.....)).)))........))))).. ( -24.80, z-score = -0.81, R) >droAna3.scaffold_13117 3673154 110 + 5790199 UCCAUUUAUUCAUAAUUUGCAAUUUUACAAAAAAAAAUAAGUAUAAUCAGUCAGGACUUCAGUCAGGAUUAGAGCA---------UCAAAGCGGAAAUGCAUUAAUCUGAACGAUUUAU .....(..((((.....((((.((((....................((((((..(((....)))..)))).))((.---------.....)))))).))))......))))..)..... ( -15.20, z-score = -0.52, R) >consensus ______CAAAUGUGCGU_GAUGUGUGCCAUAAAUGUGUCACCAUCAACAG_CGGGUUGUCAGUUAAAAUUAACGCAAUAGCGAAACUAGAGUGAAAAUCCACUCAUUUCAACAACUGAU ...............((.((((.(((.(((....))).))))))).)).........(((((((........(((....)))......(((((......)))))........))))))) (-15.03 = -16.87 + 1.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:38:17 2011