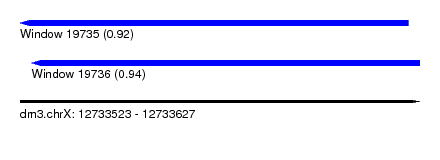
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,733,523 – 12,733,627 |
| Length | 104 |
| Max. P | 0.939987 |

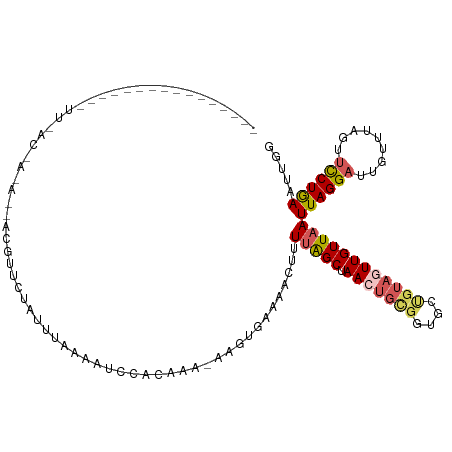
| Location | 12,733,523 – 12,733,624 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 61.32 |
| Shannon entropy | 0.64443 |
| G+C content | 0.32894 |
| Mean single sequence MFE | -20.66 |
| Consensus MFE | -8.74 |
| Energy contribution | -9.54 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.915197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
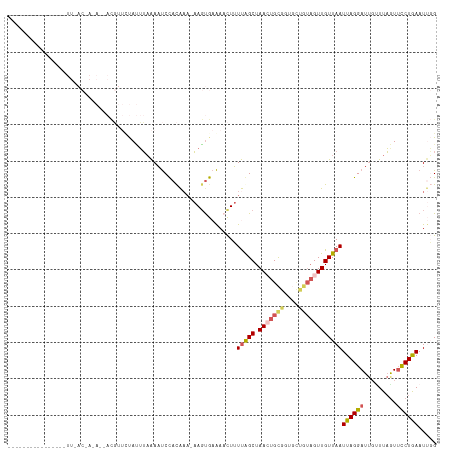
>dm3.chrX 12733523 101 - 22422827 ----------------UUUACCACAUAACGUUUUAUUUCAAAUCCACAAA-AAGUGAAAACUUUUAGCUAACUGCGGUGCUGUAGUUGUUAAUUAGGAUUGUUUAGUUCCUAAAUUGG ----------------....(((.................(((((...((-((((....))))))(((.(((((((....)))))))))).....)))))((((((...))))))))) ( -19.90, z-score = -1.21, R) >droSim1.chrX 10552388 117 - 17042790 UUUACCACAGAAAUCCUUAACAAAAGCACGUUCUAUUUAAAAUCCACAAA-AAGUGAAAACUUUUGGCUAAAUGCGGUGCUGUAGUUGUUAAUUAGGAUUGUUUAGUUUCUGAAUUGG ....(((((((((((((((((((.(((((...................((-((((....)))))).((.....)).)))))....))))))...)))))........)))))...))) ( -26.80, z-score = -2.03, R) >droSec1.super_21 484615 118 - 1102487 UUUACCACAGAAAUCCUUAACAAAAGCACGUUUUAUUUAAAAUCCACAAACAAGUGAAAACUUUUGGCUAACUGCGGUGCUGUAGUUGUUAAUUAGGAUUGCUUAGUUCCUGAAUUGG ....(((.........(((((((.(((((.................((((..(((....)))))))((.....)).)))))....)))))))((((((.........))))))..))) ( -24.80, z-score = -0.94, R) >droYak2.chrX 7010327 83 - 21770863 ---------------------------------UAGUUAGAAUCUACGA--AAUUAAAACUUUUCAGCUAACUGUGAUUUCGUAGUUGUUAAUUAGGAUUGUUUAAUGCCUGAAUCUG ---------------------------------(((((((...((((((--(((((........(((....))))))))))))))...)))))))(((((............))))). ( -16.60, z-score = -1.86, R) >droEre2.scaffold_4690 4629826 79 + 18748788 ----------------------------UUUACCACAGAAAUCCUUUACAACAGCACGACUUAUUAGCUAAAUGCG--------GUUGUUAAUUAGGAU---UUUGUUCCUGAAUCUG ----------------------------.......((((..........((((((.((..(((.....)))...))--------))))))..((((((.---.....)))))).)))) ( -15.20, z-score = -1.86, R) >consensus ________________UU_AC_A_A__ACGUUCUAUUUAAAAUCCACAAA_AAGUGAAAACUUUUAGCUAACUGCGGUGCUGUAGUUGUUAAUUAGGAUUGUUUAGUUCCUGAAUUGG ...............................................................(((((.(((((((....))))))))))))((((((.........))))))..... ( -8.74 = -9.54 + 0.80)

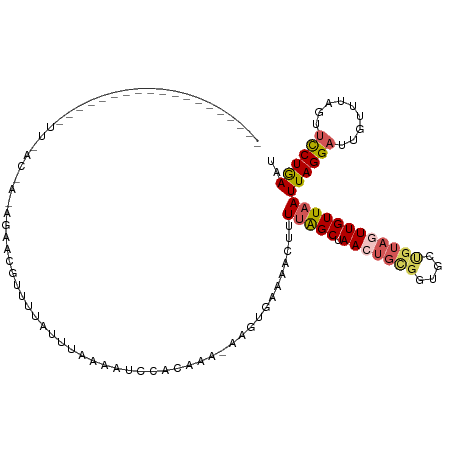
| Location | 12,733,526 – 12,733,627 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 60.43 |
| Shannon entropy | 0.66410 |
| G+C content | 0.31862 |
| Mean single sequence MFE | -19.82 |
| Consensus MFE | -8.74 |
| Energy contribution | -9.54 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.939987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12733526 101 - 22422827 ----------------CAUUUUACCACAUAACGUUUUAUUUCAAAUCCACAAA-AAGUGAAAACUUUUAGCUAACUGCGGUGCUGUAGUUGUUAAUUAGGAUUGUUUAGUUCCUAAAU ----------------...........................(((((...((-((((....))))))(((.(((((((....)))))))))).....)))))((((((...)))))) ( -18.30, z-score = -1.06, R) >droSim1.chrX 10552391 117 - 17042790 CAUUUUACCACAGAAAUCCUUAACAAAAGCACGUUCUAUUUAAAAUCCACAAA-AAGUGAAAACUUUUGGCUAAAUGCGGUGCUGUAGUUGUUAAUUAGGAUUGUUUAGUUUCUGAAU ..........((((((((((((((((.(((((...................((-((((....)))))).((.....)).)))))....))))))...)))))........)))))... ( -26.20, z-score = -2.13, R) >droSec1.super_21 484618 118 - 1102487 CAUUUUACCACAGAAAUCCUUAACAAAAGCACGUUUUAUUUAAAAUCCACAAACAAGUGAAAACUUUUGGCUAACUGCGGUGCUGUAGUUGUUAAUUAGGAUUGCUUAGUUCCUGAAU ...........((.((((((((((((.(((((.................((((..(((....)))))))((.....)).)))))....))))))...)))))).))............ ( -23.30, z-score = -0.80, R) >droYak2.chrX 7010330 83 - 21770863 ---------------------------------AGUUAGUUAGAAUCUACGA--AAUUAAAACUUUUCAGCUAACUGUGAUUUCGUAGUUGUUAAUUAGGAUUGUUUAAUGCCUGAAU ---------------------------------..((((((((...((((((--(((((........(((....))))))))))))))...))))))))................... ( -16.40, z-score = -1.61, R) >droEre2.scaffold_4690 4629829 79 + 18748788 ----------------------------GAUUUUACCACAGAAAUCCUUUACAACAGCACGACUUAUUAGCUAAAUGCG--------GUUGUUAAUUAGGAU---UUUGUUCCUGAAU ----------------------------...((((.....((((((((....((((((.((..(((.....)))...))--------))))))....)))))---))).....)))). ( -14.90, z-score = -1.86, R) >consensus ___________________UU_AC_A_AGAACGUUUUAUUUAAAAUCCACAAA_AAGUGAAAACUUUUAGCUAACUGCGGUGCUGUAGUUGUUAAUUAGGAUUGUUUAGUUCCUGAAU ..................................................................(((((.(((((((....))))))))))))((((((.........)))))).. ( -8.74 = -9.54 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:38:16 2011