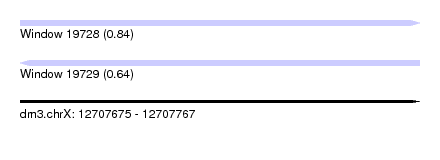
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,707,675 – 12,707,767 |
| Length | 92 |
| Max. P | 0.838182 |

| Location | 12,707,675 – 12,707,767 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 86.43 |
| Shannon entropy | 0.23609 |
| G+C content | 0.51485 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -23.36 |
| Energy contribution | -24.40 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.838182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
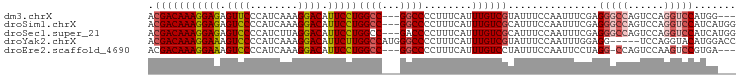
>dm3.chrX 12707675 92 + 22422827 ACGACAAAGGAGAGUUCCCAUCAAAGGACAUUCCUGGCC---GGCCCCUUUCAUUUGUCGUAUUUCCAAUUUCGAGGGCCAGUCCAGGUCCAUGG--- ((((((((.(((((...((.....(((.....)))....---))...))))).))))))))..............(((((......)))))....--- ( -27.90, z-score = -1.43, R) >droSim1.chrX 9692476 95 + 17042790 ACGACAAAGGAGAGUCCCCAUCAAAGGACAUUCCUGGCC---GGCCCCUUUCAUUUGUCGCAUUUCCAAUUUCGAGGGCCAGUCCAGGUCCAUCAUGG .(((((((((((.((((........)))).)))))((..---..))........)))))).............(((((((......))))).)).... ( -28.20, z-score = -1.23, R) >droSec1.super_21 459066 95 + 1102487 ACGACAAAGGAGAGUCCCCAUCUUAGGACAUUCCUGGCC---GACCCCUUUCAUUUGUCGCAUUUCCAAUUUCGAGGGCCAGUCCAGGUCCAUCAUGG .(((((((((((.((((........)))).)))))((..---....))......)))))).............(((((((......))))).)).... ( -27.10, z-score = -1.32, R) >droYak2.chrX 6983874 93 + 21770863 ACGACAAAGGAAAGUCCCCAUCAAAGGACAUUCUUGGCCAUGGGCCCCUUUCAUUUGUCGUAUUUCCAAUUUGGAGG-----UCCAGGUACAUGGACC ((((((((.((((((((........))))......((((...))))..)))).))))))))...(((.....)))((-----((((......)))))) ( -34.10, z-score = -2.87, R) >droEre2.scaffold_4690 4605021 91 - 18748788 ACGACAAAGGAAAGUCCCCAUCAAAGGACAUUCCUGGCC---GGCCCCUUUCAUUUGUCCUAUUUCCAAUUCCUAGG-CCAGUCCAAGUCCGUGA--- ..(((..(((((.((((........)))).)))))(((.---((((........(((.........)))......))-)).)))...))).....--- ( -22.54, z-score = -1.48, R) >consensus ACGACAAAGGAGAGUCCCCAUCAAAGGACAUUCCUGGCC___GGCCCCUUUCAUUUGUCGUAUUUCCAAUUUCGAGGGCCAGUCCAGGUCCAUGA___ .(((((((((((.((((........)))).)))))((((...))))........))))))...............(((((......)))))....... (-23.36 = -24.40 + 1.04)

| Location | 12,707,675 – 12,707,767 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 86.43 |
| Shannon entropy | 0.23609 |
| G+C content | 0.51485 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -22.60 |
| Energy contribution | -23.48 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.642574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
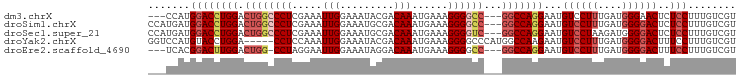
>dm3.chrX 12707675 92 - 22422827 ---CCAUGGACCUGGACUGGCCCUCGAAAUUGGAAAUACGACAAAUGAAAGGGGCC---GGCCAGGAAUGUCCUUUGAUGGGAACUCUCCUUUGUCGU ---....((((((((.((((((((.....(((.........)))......))))))---)))))))....((((.....))))....)))........ ( -31.30, z-score = -1.50, R) >droSim1.chrX 9692476 95 - 17042790 CCAUGAUGGACCUGGACUGGCCCUCGAAAUUGGAAAUGCGACAAAUGAAAGGGGCC---GGCCAGGAAUGUCCUUUGAUGGGGACUCUCCUUUGUCGU ..((((..(((((((.((((((((.....(((.........)))......))))))---)))))))...((((((....)))))).....))..)))) ( -36.60, z-score = -2.39, R) >droSec1.super_21 459066 95 - 1102487 CCAUGAUGGACCUGGACUGGCCCUCGAAAUUGGAAAUGCGACAAAUGAAAGGGGUC---GGCCAGGAAUGUCCUAAGAUGGGGACUCUCCUUUGUCGU ..((((..(((((((.((((((((.....(((.........)))......))))))---)))))))...(((((......))))).....))..)))) ( -33.40, z-score = -1.58, R) >droYak2.chrX 6983874 93 - 21770863 GGUCCAUGUACCUGGA-----CCUCCAAAUUGGAAAUACGACAAAUGAAAGGGGCCCAUGGCCAAGAAUGUCCUUUGAUGGGGACUUUCCUUUGUCGU ((((((......))))-----))(((.....)))...((((((((.(((((.((((...)))).......(((((....)))))))))).)))))))) ( -36.60, z-score = -3.11, R) >droEre2.scaffold_4690 4605021 91 + 18748788 ---UCACGGACUUGGACUGG-CCUAGGAAUUGGAAAUAGGACAAAUGAAAGGGGCC---GGCCAGGAAUGUCCUUUGAUGGGGACUUUCCUUUGUCGU ---.....(((.(((.((((-(((.....(((.........))).......)))))---)))))((((.((((((....)))))).))))...))).. ( -31.60, z-score = -1.73, R) >consensus ___UCAUGGACCUGGACUGGCCCUCGAAAUUGGAAAUACGACAAAUGAAAGGGGCC___GGCCAGGAAUGUCCUUUGAUGGGGACUCUCCUUUGUCGU .......((.((......)).))..............((((((((.......((((...)))).(((..((((((....))))))..))))))))))) (-22.60 = -23.48 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:38:10 2011