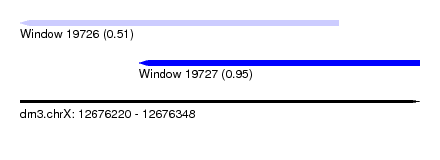
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,676,220 – 12,676,348 |
| Length | 128 |
| Max. P | 0.953530 |

| Location | 12,676,220 – 12,676,322 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 85.36 |
| Shannon entropy | 0.23501 |
| G+C content | 0.33592 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -14.99 |
| Energy contribution | -15.05 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.511100 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
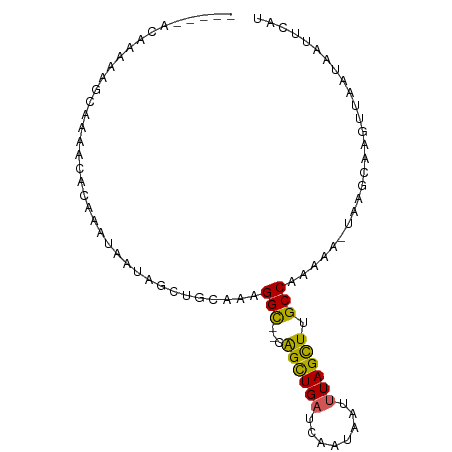
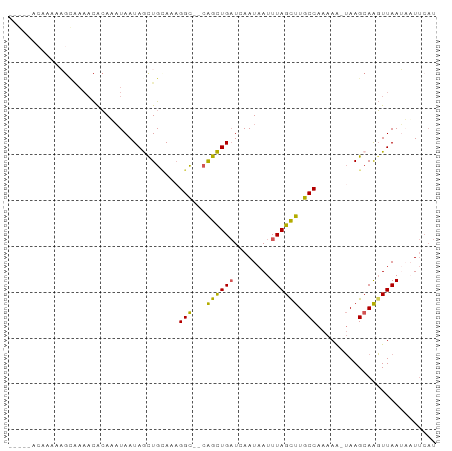
>dm3.chrX 12676220 102 - 22422827 AUAGCUGCAAAGGCCAGCUGAUCAAUAAUUUAGCUUGCCAAAAAUAAGCAAGUUAAUAAUUCAUUUCAAUGAGCAGGCUAAAUUUCGAAAAAGAAAGGAAAU .(((((((...(((.((((((........)))))).))).....................((((....))))))).))))..((((......))))...... ( -23.80, z-score = -1.89, R) >droEre2.scaffold_4690 4577723 101 + 18748788 AUAGUUGUAAAGGCAGGCUGAUCAAUAAUUUAGCUUGCCAAAAGAAAGGAGUUUAAAUUUUCAUUGCCAAAAGCAGGAGAAAUUCCCCAAAA-AAAGUCAAU ...((((....((((((((((........))))))))))........((((((.....((((.((((.....))))))))))))))......-.....)))) ( -25.70, z-score = -3.19, R) >droSec1.super_21 430761 100 - 1102487 AUAGCUGAAAAGGUCAGCUGAUCAAUAAUUUAGCUUGCCAAAAAUAAGCAAGUUAAUAAUUCAUUUCAAUGAGCAGGCUAAAUUCCCCAAA--AAAGGCAAU .(((((((.....)))))))......((((((((((((....(((......)))......((((....))))))))))))))))..((...--...)).... ( -22.20, z-score = -1.96, R) >droSim1.chrX 10510966 99 - 17042790 AUAGCUGUAAAGGCCAGCUGAUCAAUAAUUUAGCUUGCCAAAAAUAAGCAAGUUAAUAAUUCAUUUCAAUGAGCAGGCGAAAUGCCCAAA---AAAGGCAAU ....((((...(((.((((((........)))))).))).....................((((....))))))))......((((....---...)))).. ( -22.30, z-score = -1.11, R) >consensus AUAGCUGUAAAGGCCAGCUGAUCAAUAAUUUAGCUUGCCAAAAAUAAGCAAGUUAAUAAUUCAUUUCAAUGAGCAGGCGAAAUUCCCAAAA__AAAGGCAAU ....((((...(((.((((((........)))))).))).....................((((....)))))))).......................... (-14.99 = -15.05 + 0.06)

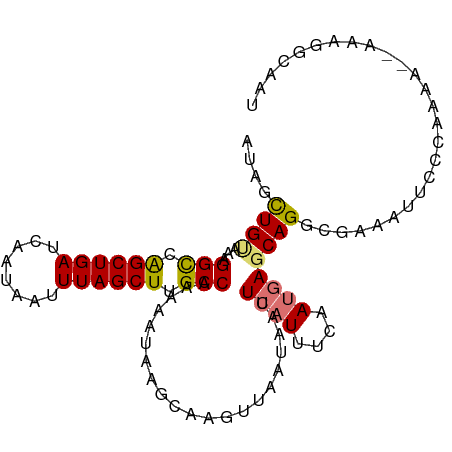
| Location | 12,676,258 – 12,676,348 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 73.55 |
| Shannon entropy | 0.46658 |
| G+C content | 0.30603 |
| Mean single sequence MFE | -16.10 |
| Consensus MFE | -9.58 |
| Energy contribution | -9.18 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.953530 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
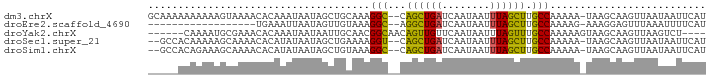
>dm3.chrX 12676258 90 - 22422827 GCAAAAAAAAAAGUAAAACACAAAUAAUAGCUGCAAAGGC--CAGCUGAUCAAUAAUUUAGCUUGCCAAAAA-UAAGCAAGUUAAUAAUUCAU ...........................(((((((....).--)))))).........(((((((((......-...)))))))))........ ( -15.40, z-score = -0.73, R) >droEre2.scaffold_4690 4577760 72 + 18748788 ------------------UGAAAUUAAUAGUUGUAAAGGC--AGGCUGAUCAAUAAUUUAGCUUGCCAAAAG-AAAGGAGUUUAAAUUUUCAU ------------------...................(((--(((((((........))))))))))....(-((((.........))))).. ( -17.90, z-score = -3.01, R) >droYak2.chrX 6953967 83 - 21770863 ------CAAAAUGCGAAACACAAAUAAUAAUUGCAACGGCAACAGUUGUUCAAUAAUUUAGUUUGCCAAAAAGUAAGCAAGUUAAGUCU---- ------......((..(((..........((((.((((((....))))))))))......((((((......))))))..)))..))..---- ( -13.30, z-score = -0.99, R) >droSec1.super_21 430797 88 - 1102487 --GCCACAAAAAGCAAAACACAUAUAAUAGCUGAAAAGGU--CAGCUGAUCAAUAAUUUAGCUUGCCAAAAA-UAAGCAAGUUAAUAAUUCAU --((........)).............(((((((.....)--)))))).........(((((((((......-...)))))))))........ ( -17.60, z-score = -2.86, R) >droSim1.chrX 10511001 88 - 17042790 --GCCACAGAAAGCAAAACACAUAUAAUAGCUGUAAAGGC--CAGCUGAUCAAUAAUUUAGCUUGCCAAAAA-UAAGCAAGUUAAUAAUUCAU --......(((................((((((.......--)))))).........(((((((((......-...)))))))))...))).. ( -16.30, z-score = -1.64, R) >consensus _____ACAAAAAGCAAAACACAAAUAAUAGCUGCAAAGGC__CAGCUGAUCAAUAAUUUAGCUUGCCAAAAA_UAAGCAAGUUAAUAAUUCAU ...........................((((((.........)))))).........(((((((((..........)))))))))........ ( -9.58 = -9.18 + -0.40)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:38:09 2011