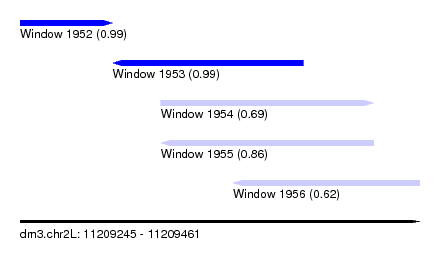
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,209,245 – 11,209,461 |
| Length | 216 |
| Max. P | 0.990922 |

| Location | 11,209,245 – 11,209,295 |
|---|---|
| Length | 50 |
| Sequences | 3 |
| Columns | 50 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Shannon entropy | 0.05510 |
| G+C content | 0.48000 |
| Mean single sequence MFE | -12.67 |
| Consensus MFE | -12.85 |
| Energy contribution | -12.63 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.10 |
| Structure conservation index | 1.01 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.985042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
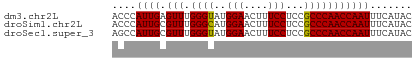
>dm3.chr2L 11209245 50 + 23011544 ACCCAUUGAGUUUGGGUAUGGAACUUUCCUCCGCCCAACCAAUUUCAUAC ....((((.(((.((((..(((....)))...)))))))))))....... ( -12.10, z-score = -1.82, R) >droSim1.chr2L 11010416 50 + 22036055 ACCCAUUGCGUUUGGGCAUGGAACUUUCCUCCGCCCAACCAAUUUCAUAC ....((((.(((.((((..(((....)))...)))))))))))....... ( -13.70, z-score = -2.74, R) >droSec1.super_3 6609779 50 + 7220098 AGCCAUUGCGUUUGGGUAUGGAACUUUCCUCCGCCCAACCAAUUUCAUAC .(..((((.(((.((((..(((....)))...)))))))))))..).... ( -12.20, z-score = -1.74, R) >consensus ACCCAUUGCGUUUGGGUAUGGAACUUUCCUCCGCCCAACCAAUUUCAUAC ....((((.(((.((((..(((....)))...)))))))))))....... (-12.85 = -12.63 + -0.22)

| Location | 11,209,295 – 11,209,398 |
|---|---|
| Length | 103 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 71.20 |
| Shannon entropy | 0.55520 |
| G+C content | 0.51841 |
| Mean single sequence MFE | -32.89 |
| Consensus MFE | -17.35 |
| Energy contribution | -17.57 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.990922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11209295 103 - 23011544 GCAGCUACAGCUUCCCAAAAAGGGAGCGCAAGUUGGUGGCAUGCGGCAGCU-AAUGUAUCUAAUAGAUACACGACAGCACGUCAAGCGCUUCCAUUGGAGCGAG----------- (((((....)))..((((...((((((((.(((((.((.....)).)))))-..((((((.....)))))).(((.....)))..)))))))).)))).))...----------- ( -36.50, z-score = -2.39, R) >droMoj3.scaffold_6500 27518543 101 - 32352404 --------AUCUCCUC-CAAUGGAA-----GUCCGAUCACAUAGAGCGGCUAAAUGUAUCUAAUAGAUACACGACAGCAUGUCGAACGCUGCCAUCCGAGCAUAAAGUCUGAGAA --------.((((.((-(...))).-----(..((.((.....))(((((....((((((.....))))))((((.....))))...)))))....))..).........)))). ( -24.60, z-score = -1.20, R) >droVir3.scaffold_12963 2294587 99 - 20206255 --------CCCUCCACACAAUGAGA-----GUCCGAUCACAUAGAGCGGCU-AAUGUAUCUAAUAGAUACACGACAGCAUGUCGAACUCUGCCAUCGGAGCAUAAA--CCGAGCG --------..(((((.....)).))-----)..............(((((.-..((((((.....))))))((((.....))))......))).((((........--)))))). ( -23.90, z-score = -1.56, R) >droWil1.scaffold_180708 4560267 102 - 12563649 ACAGAGACAGAGCGAGGGAAUGAGA----GGGGAGAAUGCAGGCAGCGGCU-AAUGUAUCUAAUAGAUACACGACAGCAUGUCAAACGCUUCCAGCGGAGCAUUAAG-------- .........................----......(((((.((((...(((-..((((((.....))))))....))).))))...(((.....)))..)))))...-------- ( -22.20, z-score = -0.51, R) >droAna3.scaffold_12916 11936252 92 + 16180835 ------GCAGUAUCU-----GGGGAUCUCUAGCCAGCGGCAUGCGGUAGCU-AAUGUAUCUAAUAGAUACACGACAGCAUGUCAAACGCUUCCAGUGGAGCGAG----------- ------((.....((-----(((....)))))...))(((((((.((....-..((((((.....))))))..)).)))))))...((((((....))))))..----------- ( -28.60, z-score = -1.06, R) >droEre2.scaffold_4929 12407285 102 + 26641161 GCAGCCGCAGCUUCC-AAAAAGGGAGCGCAAGUUGGCGGCAUGCGGCAGCU-AAUGUAUCUAAUAGAUACACGACAGCGCGUCAAGCGCUUCCAUUGGAGCGAG----------- ((.((((((((((((-.....))))))((......))....))))))..((-((((((((.....)))))..((.(((((.....))))))).))))).))...----------- ( -41.30, z-score = -2.82, R) >droYak2.chr2L 7607468 103 - 22324452 GCAGCUACAGCUUCCAAAGAAGGGAGCGCAAGUUGGCGGCAUGCGGCAGCU-AAUGUAUCUAAUAGAUACACGACAGCACGUCAAGCGCUUCCAUUGGAGCGAG----------- (((((....)))(((((....((((((((.(((((.((.....)).)))))-..((((((.....)))))).(((.....)))..)))))))).)))))))...----------- ( -39.50, z-score = -3.33, R) >droSec1.super_3 6609829 103 - 7220098 GCAGCUACAGCUUCCCAAGAAGGGAGCGCAAGUUGGCGGCAUGCGGCAGCU-AAUGUAUCUAAUAGAUACACGACAGCACGUCAAGCGCUUCCAUUGGAGCGAG----------- (((((....)))..((((...((((((((.(((((.((.....)).)))))-..((((((.....)))))).(((.....)))..)))))))).)))).))...----------- ( -39.70, z-score = -3.13, R) >droSim1.chr2L 11010466 103 - 22036055 GCAGCUACAGCUUCCCAAGAAGGGAGCGCAAGUUGGCGGCAUGCGGCAGCU-AAUGUAUCUAAUAGAUACACGACAGCACGUCAAGCGCUUCCAUUGGAGCGAG----------- (((((....)))..((((...((((((((.(((((.((.....)).)))))-..((((((.....)))))).(((.....)))..)))))))).)))).))...----------- ( -39.70, z-score = -3.13, R) >consensus GCAGCUACAGCUUCCCAAAAAGGGAGCGCAAGUUGGCGGCAUGCGGCAGCU_AAUGUAUCUAAUAGAUACACGACAGCACGUCAAGCGCUUCCAUUGGAGCGAG___________ .........((((.................(((((.((.....)).)))))...((((((.....)))))).(((.....)))))))(((((....))))).............. (-17.35 = -17.57 + 0.21)

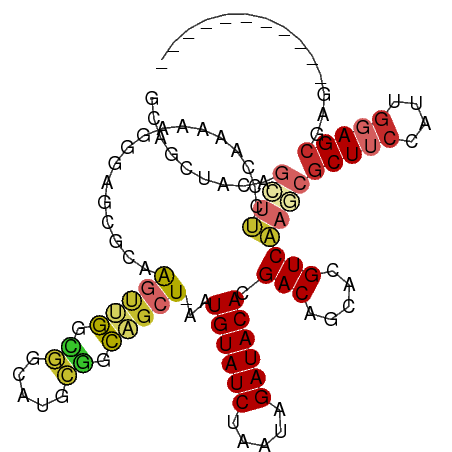

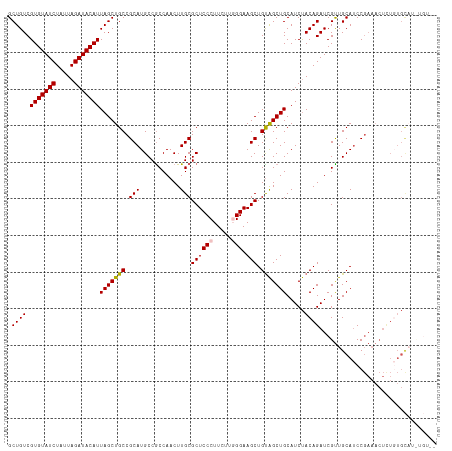
| Location | 11,209,321 – 11,209,436 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 87.56 |
| Shannon entropy | 0.21882 |
| G+C content | 0.50307 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -26.58 |
| Energy contribution | -26.66 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.686929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11209321 115 + 23011544 GCUGUCGUGUAUCUAUUAGAUACAUUAGCUGCCGCAUGCCACCAACUUGCGCUCCCUUUUUGGGAAGCUGUAGCUGCAUCUACAGAUCGUUGCAUCCGAAACUCUGUGCAUUUGU-- ((....(((((((.....))))))).((((((.((..((((......)).))((((.....)))).)).))))))))...(((((((((.......)))...)))))).......-- ( -32.40, z-score = -1.27, R) >droEre2.scaffold_4929 12407311 96 - 26641161 GCUGUCGUGUAUCUAUUAGAUACAUUAGCUGCCGCAUGCCGCCAACUUGCGCUCCCUUUUU-GGAAGCUGCGGCUGCAUCCACAGAUCGCUGCAUCC-------------------- ((.((((((((((.....)))))))..((.((((((....((......))(((.((.....-)).))))))))).)).......))).)).......-------------------- ( -32.60, z-score = -2.26, R) >droYak2.chr2L 7607494 109 + 22324452 GCUGUCGUGUAUCUAUUAGAUACAUUAGCUGCCGCAUGCCGCCAACUUGCGCUCCCUUCUUUGGAAGCUGUAGCUGCAUCUACAGAUCGUUGCAACCGAAACUCUCUGU-------- ((((..(((((((.....)))))))))))(((.((.(((.((......))(((.((......)).))).))))).)))...(((((..(((........)))..)))))-------- ( -29.40, z-score = -1.36, R) >droSec1.super_3 6609855 117 + 7220098 GCUGUCGUGUAUCUAUUAGAUACAUUAGCUGCCGCAUGCCGCCAACUUGCGCUCCCUUCUUGGGAAGCUGUAGCUGCAUCUACAGAUCGUUGCAUCCGAAACUUUGUGCAUCUGUUG ((....(((((((.....))))))).((((((.((..((.((......))))((((.....)))).)).))))))))....(((((....(((((..........)))))))))).. ( -34.30, z-score = -1.49, R) >droSim1.chr2L 11010492 117 + 22036055 GCUGUCGUGUAUCUAUUAGAUACAUUAGCUGCCGCAUGCCGCCAACUUGCGCUCCCUUCUUGGGAAGCUGUAGCUGCAUCUACAGAUCGUUGCAUCCGAAACUCUGUGCAUCUGUUG ((....(((((((.....))))))).((((((.((..((.((......))))((((.....)))).)).))))))))....(((((....(((((..........)))))))))).. ( -34.30, z-score = -1.42, R) >consensus GCUGUCGUGUAUCUAUUAGAUACAUUAGCUGCCGCAUGCCGCCAACUUGCGCUCCCUUCUUGGGAAGCUGUAGCUGCAUCUACAGAUCGUUGCAUCCGAAACUCUGUGCAU_UGU__ .((((.(((((((.....)))))))(((((((.(((...........)))((((((.....))).))).))))))).....))))................................ (-26.58 = -26.66 + 0.08)


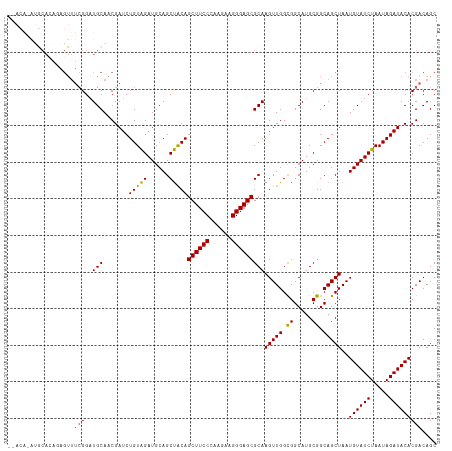
| Location | 11,209,321 – 11,209,436 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 87.56 |
| Shannon entropy | 0.21882 |
| G+C content | 0.50307 |
| Mean single sequence MFE | -34.96 |
| Consensus MFE | -30.92 |
| Energy contribution | -30.68 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.856590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11209321 115 - 23011544 --ACAAAUGCACAGAGUUUCGGAUGCAACGAUCUGUAGAUGCAGCUACAGCUUCCCAAAAAGGGAGCGCAAGUUGGUGGCAUGCGGCAGCUAAUGUAUCUAAUAGAUACACGACAGC --.....((((((((...(((.......))))))))...(((((((((....((((.....))))((....))..))))).)))))))(((..((((((.....))))))....))) ( -35.10, z-score = -1.61, R) >droEre2.scaffold_4929 12407311 96 + 26641161 --------------------GGAUGCAGCGAUCUGUGGAUGCAGCCGCAGCUUCC-AAAAAGGGAGCGCAAGUUGGCGGCAUGCGGCAGCUAAUGUAUCUAAUAGAUACACGACAGC --------------------............((((.(..((.((((((((((((-.....))))))((......))....)))))).))...((((((.....))))))).)))). ( -35.30, z-score = -1.91, R) >droYak2.chr2L 7607494 109 - 22324452 --------ACAGAGAGUUUCGGUUGCAACGAUCUGUAGAUGCAGCUACAGCUUCCAAAGAAGGGAGCGCAAGUUGGCGGCAUGCGGCAGCUAAUGUAUCUAAUAGAUACACGACAGC --------.............((((.....(((((((((((((......((((((......))))))...(((((.((.....)).)))))..))))))).))))))...))))... ( -34.10, z-score = -1.61, R) >droSec1.super_3 6609855 117 - 7220098 CAACAGAUGCACAAAGUUUCGGAUGCAACGAUCUGUAGAUGCAGCUACAGCUUCCCAAGAAGGGAGCGCAAGUUGGCGGCAUGCGGCAGCUAAUGUAUCUAAUAGAUACACGACAGC ....(((((((...(((((((.((((..(.(((....))).(((((...((((((......))))))...))))))..)))).))).))))..)))))))................. ( -34.60, z-score = -1.31, R) >droSim1.chr2L 11010492 117 - 22036055 CAACAGAUGCACAGAGUUUCGGAUGCAACGAUCUGUAGAUGCAGCUACAGCUUCCCAAGAAGGGAGCGCAAGUUGGCGGCAUGCGGCAGCUAAUGUAUCUAAUAGAUACACGACAGC ....(((((((...(((((((.((((..(.(((....))).(((((...((((((......))))))...))))))..)))).))).))))..)))))))................. ( -35.70, z-score = -1.46, R) >consensus __ACA_AUGCACAGAGUUUCGGAUGCAACGAUCUGUAGAUGCAGCUACAGCUUCCCAAGAAGGGAGCGCAAGUUGGCGGCAUGCGGCAGCUAAUGUAUCUAAUAGAUACACGACAGC ..................(((..(((.......(((((......)))))((((((......)))))))))(((((.((.....)).)))))..((((((.....))))))))).... (-30.92 = -30.68 + -0.24)


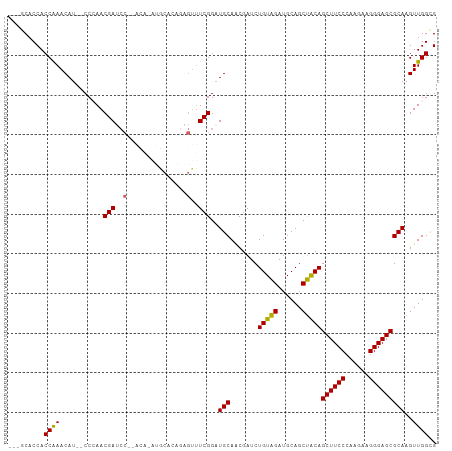
| Location | 11,209,360 – 11,209,461 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.18 |
| Shannon entropy | 0.30414 |
| G+C content | 0.53325 |
| Mean single sequence MFE | -26.33 |
| Consensus MFE | -21.00 |
| Energy contribution | -20.36 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.622474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11209360 101 - 23011544 ---GCACCACCAAACAU--CCCAACGAUCC--ACAAAUGCACAGAGUUUCGGAUGCAACGAUCUGUAGAUGCAGCUACAGCUUCCCAAAAAGGGAGCGCAAGUUGGUG ---....((((((..((--((.(((.....--.............)))..))))((.......(((((......)))))((((((......))))))))...)))))) ( -25.17, z-score = -1.00, R) >droEre2.scaffold_4929 12407350 81 + 26641161 ---GCACCACCGAACAU--CCCAACGA---------------------UCGGAUGCAGCGAUCUGUGGAUGCAGCCGCAGCUUCCAAA-AAGGGAGCGCAAGUUGGCG ---(((((((((..(((--((......---------------------..)))))...))....)))).))).(((((.((((((...-..)))))))).....))). ( -29.10, z-score = -1.75, R) >droYak2.chr2L 7607533 98 - 22324452 GCACCACCACCAAACAU--CCCAACGAUCC--------ACAGAGAGUUUCGGUUGCAACGAUCUGUAGAUGCAGCUACAGCUUCCAAAGAAGGGAGCGCAAGUUGGCG ((...............--.......(((.--------(((((..(((........)))..))))).))).(((((...((((((......))))))...))))))). ( -26.70, z-score = -1.60, R) >droSec1.super_3 6609894 105 - 7220098 ---GCACCACCAAACAUCACCAAACGAUCCCAACAGAUGCACAAAGUUUCGGAUGCAACGAUCUGUAGAUGCAGCUACAGCUUCCCAAGAAGGGAGCGCAAGUUGGCG ---..........................(((((...(((....((((.((..((((......))))..)).))))...((((((......))))))))).))))).. ( -24.20, z-score = -0.65, R) >droSim1.chr2L 11010531 105 - 22036055 ---GCACCACCAAACAUCACCAAACGAUCCCAACAGAUGCACAGAGUUUCGGAUGCAACGAUCUGUAGAUGCAGCUACAGCUUCCCAAGAAGGGAGCGCAAGUUGGCG ---((.........((((........(((......)))..(((((...(((.......)))))))).))))(((((...((((((......))))))...))))))). ( -26.50, z-score = -1.18, R) >consensus ___GCACCACCAAACAU__CCCAACGAUCC__ACA_AUGCACAGAGUUUCGGAUGCAACGAUCUGUAGAUGCAGCUACAGCUUCCCAAGAAGGGAGCGCAAGUUGGCG .........((((...........(((.....................)))..(((.......(((((......)))))((((((......)))))))))..)))).. (-21.00 = -20.36 + -0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:51 2011