
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,664,489 – 12,664,589 |
| Length | 100 |
| Max. P | 0.597924 |

| Location | 12,664,489 – 12,664,589 |
|---|---|
| Length | 100 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 53.91 |
| Shannon entropy | 0.96302 |
| G+C content | 0.65893 |
| Mean single sequence MFE | -44.84 |
| Consensus MFE | -11.78 |
| Energy contribution | -10.73 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 2.23 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.597924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
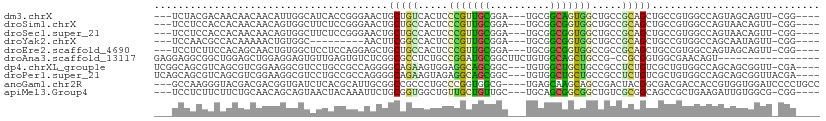
>dm3.chrX 12664489 100 + 22422827 ---UCUACGACAACAACAACAUUGGCAUCACCGGGAACUGCUGUCACUCCCGUUGCGGA---UGCGGCAGUGGCUGCCGCAGCUGCCGUGGCCAGUAGCAGUU-CGG---- ---..................((((.....))))((((((((...(((...((..(((.---((((((((...))))))))....)))..)).))))))))))-)..---- ( -38.30, z-score = -0.71, R) >droSim1.chrX 9666794 100 + 17042790 ---UCCUCCACCACAACAACAGUGGCUUCUCCGGGAACUGCUGCCACUCCCGUUGCGGA---UGCGGCGGUGGCUGCCGCAGCUGCCGUGGCCAGUAACAGUU-CGG---- ---....((.((((.......)))).........(((((((((((((....(((((...---.)))))((..((((...))))..)))))).)))...)))))-)))---- ( -38.30, z-score = -0.11, R) >droSec1.super_21 419936 100 + 1102487 ---UCCUCCACCACAACAACAGUGGCUUCUCCGGGAACUGCUGCCACUCCCGUUGCGGA---UGCGGCGGUGGCUGCCGCAGCUGCCGUGGCCAGUAACAGUU-CGG---- ---....((.((((.......)))).........(((((((((((((....(((((...---.)))))((..((((...))))..)))))).)))...)))))-)))---- ( -38.30, z-score = -0.11, R) >droYak2.chrX 6946099 91 + 21770863 ---UCCAACGCCACAAAAACUGUGGC---------AACUUCGGCCACUCCCGUUGCGGA---UGCGGCGGUGGCUGCCGCAGCUGCCGUGGCCAGCAAUAGUU-CGG---- ---.(((((((((((.....))))))---------......((((((....(((((...---.)))))((..((((...))))..)))))))).......)))-.))---- ( -42.20, z-score = -2.31, R) >droEre2.scaffold_4690 4566613 100 - 18748788 ---UCCUCUUCCACAGCAACUGUGGCUCCUCCAGGAGCUGCUGCCACUCCCGUUGCGGA---UGCGGCGGUGGCCGCCGCAGCUGCCGUGGCCAGUAGCAGUU-CGG---- ---.((....((((((...)))))).........((((((((((.....(((((((...---.)))))))(((((((.((....)).))))))))))))))))-)))---- ( -49.30, z-score = -2.16, R) >droAna3.scaffold_13117 3590870 93 + 5790199 GAGGAGGCGGCUGGAGCUGGAGGAGUGUUGAGUGUCUCGGCGCCUCUGCCGGAUGCGGCUUCUGUGGCAGCUGCCG-CCGCGGUGGCGAACAGU----------------- ..((.((((((((..((.((((((((((((((...))))))).))))((((....))))))).))..)))))))).-))((....)).......----------------- ( -41.00, z-score = 0.14, R) >dp4.chrXL_group1e 6540326 103 - 12523060 UCGGCAGCGUCAGCGUCGGAAGGCGUCCUGCCGCCAGGGGCAGAAGUGGAGGCAGCGGC---UGUGGCUGCUGCCGCCUCUGUCGCUGUGGCCAGCAGCGGUU-CGA---- ..(((((.(...(((((....)))))))))))(((...((((((...((.(((((((((---....))))))))).))))))))(((((.....)))))))).-...---- ( -57.00, z-score = -1.76, R) >droPer1.super_21 704361 104 - 1645244 UCAGCAGCGUCAGCGUCGGAAGGCGUCCUGCCGCCAGGGGCAGAAGUAGAGGCAGCGGC---UGUGGCUGCUGCCGCCUCUGUCGCUGUGGCCAGCAGCGGUUACGA---- ...((.((..(((((.((((.(((...(((((......))))).......(((((((((---....)))))))))))))))).)))))..))..))..((....)).---- ( -52.80, z-score = -1.10, R) >anoGam1.chr2R 14021409 104 + 62725911 ---GCCAAGGGUACGACGACGGUGAUCUCACGCAUUGCGGCCGCCCUGCCCGGUGGCG----UGAGCAAGCAGCCGACUACGGCGACGACCACCGUGGUGGAUCCCCUGCC ---((..((((..(.((.((((((.((((..((.((((.((((((......)))))).----...)))))).((((....)))))).)).)))))).)).)...)))))). ( -46.90, z-score = -1.46, R) >apiMel3.Group4 2215881 100 + 10796202 ---UCCUCUUCUUCUGCAACAGCAGUAACUACAAAUUCUGCGGUGGCUGUUGCUGUUGC---UGCAGCGGCGGCUGUCGCGGCAGCCGCUGAAGAUUGUGGCG-CGG---- ---..........((((....................((((((..(((((((((((...---.)))))))))))..))))))..(((((........))))))-)))---- ( -44.30, z-score = -1.96, R) >consensus ___UCCUCGUCCACAACAACAGUGGCCUCUCCGGGAACUGCUGCCACUCCCGUUGCGGA___UGCGGCGGCGGCCGCCGCAGCUGCCGUGGCCAGUAGCAGUU_CGG____ .......................................(((((.....(((((((.(......).))))))).....)))))............................ (-11.78 = -10.73 + -1.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:38:06 2011