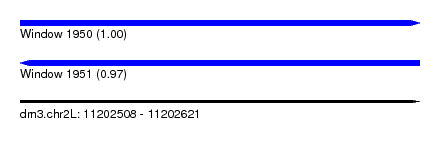
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 11,202,508 – 11,202,621 |
| Length | 113 |
| Max. P | 0.997275 |

| Location | 11,202,508 – 11,202,621 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 139 |
| Reading direction | forward |
| Mean pairwise identity | 66.49 |
| Shannon entropy | 0.58105 |
| G+C content | 0.46890 |
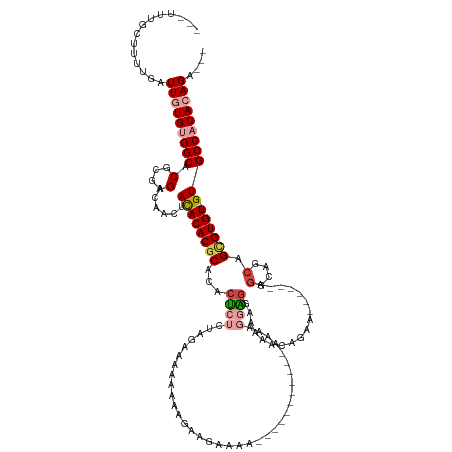
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -18.31 |
| Energy contribution | -18.75 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.997275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
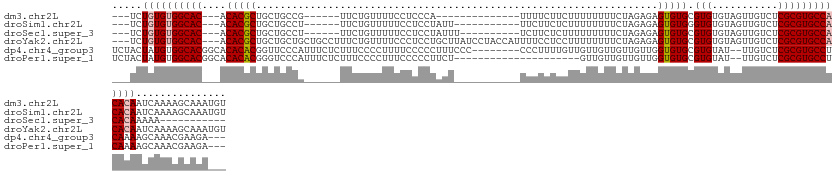
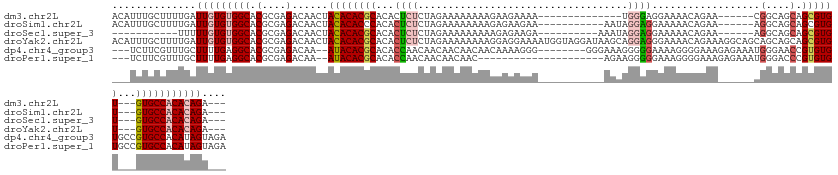
>dm3.chr2L 11202508 113 + 23011544 ---UCUGUGUGGCAC---ACACGCUGCUGCCG------UUCUGUUUUCCUCCCA--------------UUUUCUUCUUUUUUUUCUAGAGAGUGUGCGUGUGUAGUUGUCUCGCGUGCCACACAAUCAAAAGCAAAUGU ---..((((((((((---((((((.((....)------).............((--------------((((((............)))))))).))))))((((....)).))))))))))))............... ( -31.90, z-score = -2.47, R) >droSim1.chr2L 11003659 116 + 22036055 ---UCUGUGUGGCAC---ACACGCUGCUGCCU------UUCUGUUUUUCCUCCUAUU-----------UUCUUCUCUUUUUUUUCUAGAGAGUGUGGGUGUGUAGUUGUCUCGCGUGCCACACAAUCAAAAGCAAAUGU ---..((((((((((---((((((..(..(((------(((((..............-----------.................))))))).)..))))))).((......))))))))))))............... ( -32.68, z-score = -2.74, R) >droSec1.super_3 6603128 106 + 7220098 ---UCUGUGUGGCAC---ACACGCUGCUGCCU------UUCUGUUUUUCCUCCUAUUU----------UCUUCUCUUUUUUUUUCUAGAGAGUGUGCGUGUGUAGUUGUCUCGCGUGCCACACAAAAA----------- ---..((((((((((---((((((.((.((((------(((((...............----------.................))))))).)))))))))).((......))))))))))))....----------- ( -33.15, z-score = -4.71, R) >droYak2.chr2L 7600562 133 + 22324452 ---UCUGUGUGGCAC---ACACGCUGCUGCUGCUGCCUUUCUGUUUUCCCUCCUGCUUAUCCUACCAUUUUCCUCCUUUUUUUUCUAGAGAGUGUGCGUGUGUAGUUGUCUCGCGUGCCACACAAUCAAAAGCAAAUGU ---..((((((((((---((((((.((....)).(((((((((..........................................))))))).))))))))((((....)).))))))))))))............... ( -33.45, z-score = -2.02, R) >dp4.chr4_group3 6577253 126 + 11692001 UCUACUAUGUGGCACGGCACACACGGUUCCCAUUUCUCUUUCCCCUUUUCCCCCUUUCCC--------CCCUUUUGUUGUUGUUGUUGUUGGUGUGCGUGUAU--UUGUCUCGCGUGCCUCAAAAGCAAACGAAGA--- ...((..((((((...)).))))..)).................................--------...(((((((..((((.(((..((..((((.(...--....).))))..)).))).))))))))))).--- ( -25.60, z-score = -0.49, R) >droPer1.super_1 8067996 113 + 10282868 UCUACUAUGUGGCACGGCACACACGGGUCCCAUUUCUCUUUCCCCUUUCCCCCUUCU---------------------GUUGUUGUUGUUGGUGUGCGUGUAU--UUGUCUCGCGUGCCUCAAAAGCAAACGAAGA--- ....(..((((((...)).))))..)..........................((((.---------------------.(((((.(((..((..((((.(...--....).))))..)).))).)))))..)))).--- ( -27.40, z-score = -0.42, R) >consensus ___UCUGUGUGGCAC___ACACGCUGCUGCCA______UUCUGUUUUUCCCCCUAUU___________UUUUCUUCUUUUUUUUCUAGAGAGUGUGCGUGUGUAGUUGUCUCGCGUGCCACACAAUCAAAAGCAAA___ .....((((((((((....(((((...................................................................))))).(((...........)))))))))))))............... (-18.31 = -18.75 + 0.44)
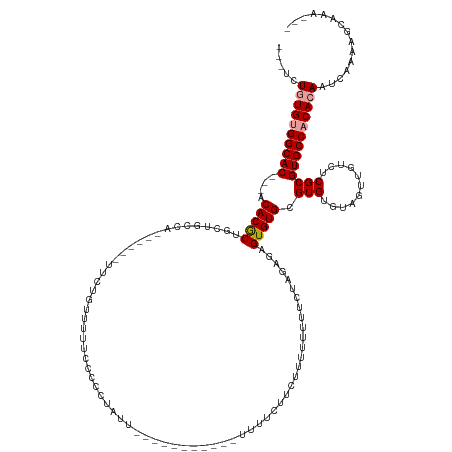
| Location | 11,202,508 – 11,202,621 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 139 |
| Reading direction | reverse |
| Mean pairwise identity | 66.49 |
| Shannon entropy | 0.58105 |
| G+C content | 0.46890 |
| Mean single sequence MFE | -31.41 |
| Consensus MFE | -17.51 |
| Energy contribution | -18.17 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 11202508 113 - 23011544 ACAUUUGCUUUUGAUUGUGUGGCACGCGAGACAACUACACACGCACACUCUCUAGAAAAAAAAGAAGAAAA--------------UGGGAGGAAAACAGAA------CGGCAGCAGCGUGU---GUGCCACACAGA--- ..............(((((((((.(....)......((((((((...(((((...................--------------.)))))..........------.(....).))))))---))))))))))).--- ( -33.15, z-score = -2.32, R) >droSim1.chr2L 11003659 116 - 22036055 ACAUUUGCUUUUGAUUGUGUGGCACGCGAGACAACUACACACCCACACUCUCUAGAAAAAAAAGAGAAGAA-----------AAUAGGAGGAAAAACAGAA------AGGCAGCAGCGUGU---GUGCCACACAGA--- ..............((((((((((((((...........(..((....(((((.........)))))....-----------....))..)..........------..((....)).)))---))))))))))).--- ( -28.70, z-score = -1.51, R) >droSec1.super_3 6603128 106 - 7220098 -----------UUUUUGUGUGGCACGCGAGACAACUACACACGCACACUCUCUAGAAAAAAAAAGAGAAGA----------AAAUAGGAGGAAAAACAGAA------AGGCAGCAGCGUGU---GUGCCACACAGA--- -----------..((((((((((.(....)......((((((((....(((((..........)))))...----------........(......)....------........))))))---))))))))))))--- ( -33.40, z-score = -4.31, R) >droYak2.chr2L 7600562 133 - 22324452 ACAUUUGCUUUUGAUUGUGUGGCACGCGAGACAACUACACACGCACACUCUCUAGAAAAAAAAGGAGGAAAAUGGUAGGAUAAGCAGGAGGGAAAACAGAAAGGCAGCAGCAGCAGCGUGU---GUGCCACACAGA--- ..............(((((((((.(....)......((((((((...((((((.................................))))))..............((....)).))))))---))))))))))).--- ( -36.41, z-score = -1.48, R) >dp4.chr4_group3 6577253 126 - 11692001 ---UCUUCGUUUGCUUUUGAGGCACGCGAGACAA--AUACACGCACACCAACAACAACAACAACAAAAGGG--------GGGAAAGGGGGAAAAGGGGAAAGAGAAAUGGGAACCGUGUGUGCCGUGCCACAUAGUAGA ---......((((((..((.((((((.(......--...(((((((.((..(.....(..(.......)..--------).....)..))....((.................)))))))))))))))).)).)))))) ( -27.03, z-score = 0.70, R) >droPer1.super_1 8067996 113 - 10282868 ---UCUUCGUUUGCUUUUGAGGCACGCGAGACAA--AUACACGCACACCAACAACAACAAC---------------------AGAAGGGGGAAAGGGGAAAGAGAAAUGGGACCCGUGUGUGCCGUGCCACAUAGUAGA ---......((((((..((.((((((.(......--...(((((((.((..(..(......---------------------.)..)..))...(((...............))))))))))))))))).)).)))))) ( -29.76, z-score = -0.49, R) >consensus ___UUUGCUUUUGAUUGUGUGGCACGCGAGACAACUACACACGCACACUCUCUAGAAAAAAAAGAAGAAAA___________AAAAGGAGGAAAAACAGAA______AGGCAGCAGCGUGU___GUGCCACACAGA___ ..............((((((((((((.....)......((((((.......................................................................))))))...))))))))))).... (-17.51 = -18.17 + 0.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:31:47 2011