
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,568,451 – 12,568,543 |
| Length | 92 |
| Max. P | 0.892046 |

| Location | 12,568,451 – 12,568,543 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 69.37 |
| Shannon entropy | 0.53618 |
| G+C content | 0.39215 |
| Mean single sequence MFE | -19.48 |
| Consensus MFE | -9.91 |
| Energy contribution | -10.25 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.892046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12568451 92 - 22422827 UAAACGGCAAAGGAGAGACGUCUUUUUCUUUCCUUUAAAAACGUGUUCGAGUUCG------------AAAAACAACUUUAAGACAAUCUACCUCACUUCACGCC------- .....(((((((((((((........))))))))))......((.((.(((((..------------......))))).)).)).................)))------- ( -20.30, z-score = -1.85, R) >droSim1.chrX_random 3478970 91 - 5698898 UAAACGGCAAAGGAAAGACGUCUUUUUCUUUCCUUU-AAAACGUGUUCGAGUUCG------------AGAAACAACUUUAAAACAAUCUACCUCACUUCACGCC------- .....(((((((((((((........))))))))))-.....(((...(((....------------(((................)))..)))....))))))------- ( -19.39, z-score = -2.07, R) >droSec1.super_21 322527 91 - 1102487 UAAACGGCAAAGGAAAGACGUCUUUUUCUUUCCUUU-AAAACGUGUUCGAGUUCG------------AGAAACAACUUUAAGACAAUCUACCUCACUUCACGCC------- .....(((((((((((((........))))))))))-.....((.((.(((((..------------......))))).)).)).................)))------- ( -20.20, z-score = -1.85, R) >droYak2.chrX 6840131 109 - 21770863 UAAACGGC-AAGGAAAGACGUCUUUUUCUUUCCCUU-UAAACGUGUUCGAGUUCGAAAAAACAAAAAAAAAACAACUUUAAGACAAUCCCCCUUUCCACACACUUCACGCC .....(((-..(((((((........)))))))...-.....((.((((....))))...))..............................................))) ( -16.50, z-score = -0.79, R) >droEre2.scaffold_4690 4458270 92 + 18748788 UAAACGGC-AAGGAAAGACGUCUUUUUCUUUCCUUU-UAAACGUGUUCGAGUUCG------------AAAAACAACUUUAAGACAAUCUACCUCACUUCACGCCGU----- ...(((((-(((((((((........))))))))).-.....((.((.(((((..------------......))))).)).)).................)))))----- ( -22.80, z-score = -2.79, R) >droPer1.super_77 117093 84 + 256778 UAGAUCGCAGUAGAGUCUGGUGUUUUGGAUACCCUU---UAUUUGUCCCUUUUCCA-------GGGUGUAUAUGGACUUCGAACAUUGCAUUUU----------------- ......(((((.((((((((((((...))))))...---.......((((.....)-------))).......)))))).....))))).....----------------- ( -17.70, z-score = -0.14, R) >consensus UAAACGGCAAAGGAAAGACGUCUUUUUCUUUCCUUU_AAAACGUGUUCGAGUUCG____________AAAAACAACUUUAAGACAAUCUACCUCACUUCACGCC_______ .....(((((((((((((........))))))))))......((.((.(((((....................))))).)).)).................)))....... ( -9.91 = -10.25 + 0.34)
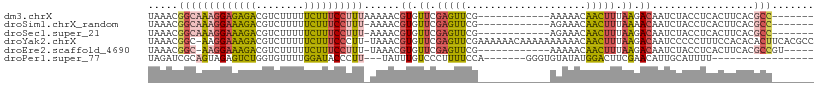
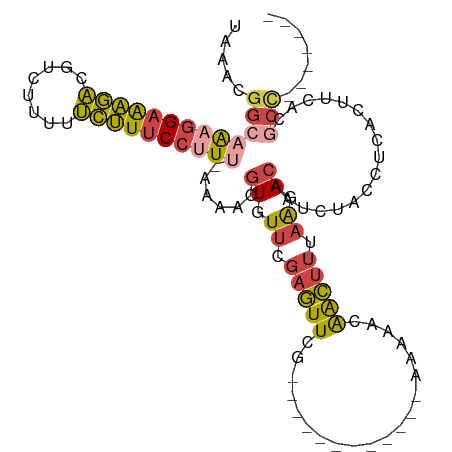

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:37:57 2011