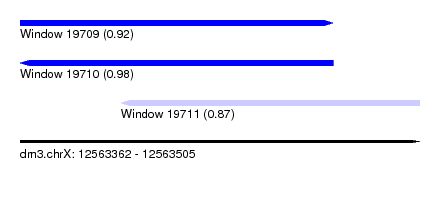
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,563,362 – 12,563,505 |
| Length | 143 |
| Max. P | 0.977413 |

| Location | 12,563,362 – 12,563,474 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.70 |
| Shannon entropy | 0.20149 |
| G+C content | 0.31441 |
| Mean single sequence MFE | -26.26 |
| Consensus MFE | -19.16 |
| Energy contribution | -18.88 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.916028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
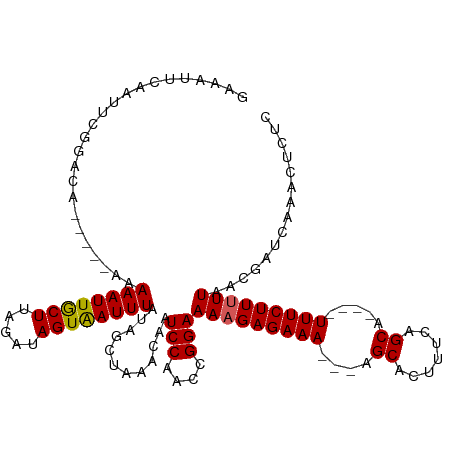
>dm3.chrX 12563362 112 + 22422827 ----GAAAUAGGUAAUUGAAUCCAGCAAAAUAUGUAAUUGGAAAGUUUGAUCGUUAAAAAGAAA----UGCUGAAAGUGCUGCUUUUCUCUUUUCCGGUUGGAUUGUUUAGAUAUAAAUU ----.......(((.((((((((((((.....)))...)))).((((..((((..(((.(((((----.((..........)).))))).)))..))))..)))).))))).)))..... ( -20.80, z-score = -0.77, R) >droSim1.chrX_random 3473652 113 + 5698898 GAAGUAGGUAUGUAAUUCAAUCCGGCAAAAUAUGUAAUUGGAGAGUUUGAUUGAUAAAAAGAAA----UGCUGAAAGUGCU---UUUCUCUUUUCCGGUUGGAUUGUUUAGCUAUAAAUU ...((((.((..((((((((.((((.((((....((((..((...))..))))......(((((----.((.......)).---))))).))))))))))))))))..)).))))..... ( -25.70, z-score = -1.96, R) >droSec1.super_21 317523 117 + 1102487 GAAAUAGGUAUGUAAUUCAAUCCGGCAAAAUAUGUAAUUGGAGAGUUUGAUUGAUAAAAAGAAAGAAAUGCUGAAAGUGCU---UUUCUCUUUUCCGGUUGGAUUGUUUAGCUAUAAAUU ...((((.((..((((((((.((((.........((((..((...))..))))...((((((..((((.((.......)).---))))))))))))))))))))))..)).))))..... ( -26.20, z-score = -2.10, R) >droYak2.chrX 6835291 109 + 21770863 ----AAAAUAGGUAAUUCAAUCCAGCAAAAUAUGUAAUUGGAGAGUUUGAUCGUUAAAAAGAAA----UGCAGAAAGUGCA---UUUCUCUUUUCCGGUUGGAUUGUUUGGCUAUAAAUC ----...((((.(((..(((((((((..........((..((...))..))((..(((.(((((----((((.....))))---))))).)))..))))))))))).))).))))..... ( -30.50, z-score = -4.13, R) >droEre2.scaffold_4690 4453477 109 - 18748788 ----GAAAUAUGUAAUUCAAUCCAUCAAAAUAUGUAAUUGGAGAGUUUGAUCGUUAAAAAGAAA----UGCAGAAAGUGCA---UUUCUCGUUUCCGGUUGGAUUGUUUGGCUAUAAAUC ----............................((((.(..((.((((..((((..((..(((((----((((.....))))---)))))..))..))))..)))).))..).)))).... ( -28.10, z-score = -3.43, R) >consensus ____GAAAUAUGUAAUUCAAUCCAGCAAAAUAUGUAAUUGGAGAGUUUGAUCGUUAAAAAGAAA____UGCUGAAAGUGCU___UUUCUCUUUUCCGGUUGGAUUGUUUAGCUAUAAAUU ................................((((.(((((.((((..((((..(((.(((((.....((.......))....))))).)))..))))..)))).))))).)))).... (-19.16 = -18.88 + -0.28)


| Location | 12,563,362 – 12,563,474 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.70 |
| Shannon entropy | 0.20149 |
| G+C content | 0.31441 |
| Mean single sequence MFE | -19.10 |
| Consensus MFE | -13.72 |
| Energy contribution | -14.12 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12563362 112 - 22422827 AAUUUAUAUCUAAACAAUCCAACCGGAAAAGAGAAAAGCAGCACUUUCAGCA----UUUCUUUUUAACGAUCAAACUUUCCAAUUACAUAUUUUGCUGGAUUCAAUUACCUAUUUC---- ...............((((((..((..(((((((((.((..........)).----)))))))))..)).................((.....)).))))))..............---- ( -15.00, z-score = -1.63, R) >droSim1.chrX_random 3473652 113 - 5698898 AAUUUAUAGCUAAACAAUCCAACCGGAAAAGAGAAA---AGCACUUUCAGCA----UUUCUUUUUAUCAAUCAAACUCUCCAAUUACAUAUUUUGCCGGAUUGAAUUACAUACCUACUUC ..............((((((....((.(((((((((---.((.......)).----))))))))).))..................((.....))..))))))................. ( -17.00, z-score = -2.53, R) >droSec1.super_21 317523 117 - 1102487 AAUUUAUAGCUAAACAAUCCAACCGGAAAAGAGAAA---AGCACUUUCAGCAUUUCUUUCUUUUUAUCAAUCAAACUCUCCAAUUACAUAUUUUGCCGGAUUGAAUUACAUACCUAUUUC ..............((((((.....(((((((((((---.((.......)).))))..))))))).....................((.....))..))))))................. ( -15.20, z-score = -1.62, R) >droYak2.chrX 6835291 109 - 21770863 GAUUUAUAGCCAAACAAUCCAACCGGAAAAGAGAAA---UGCACUUUCUGCA----UUUCUUUUUAACGAUCAAACUCUCCAAUUACAUAUUUUGCUGGAUUGAAUUACCUAUUUU---- ..............(((((((..((..(((((((((---((((.....))))----)))))))))..)).................((.....)).))))))).............---- ( -24.80, z-score = -4.74, R) >droEre2.scaffold_4690 4453477 109 + 18748788 GAUUUAUAGCCAAACAAUCCAACCGGAAACGAGAAA---UGCACUUUCUGCA----UUUCUUUUUAACGAUCAAACUCUCCAAUUACAUAUUUUGAUGGAUUGAAUUACAUAUUUC---- ..............((((((...((..((.((((((---((((.....))))----)))))).))..))((((((................)))))))))))).............---- ( -23.49, z-score = -4.31, R) >consensus AAUUUAUAGCUAAACAAUCCAACCGGAAAAGAGAAA___AGCACUUUCAGCA____UUUCUUUUUAACGAUCAAACUCUCCAAUUACAUAUUUUGCUGGAUUGAAUUACAUAUUUA____ ..............((((((....(..(((((((((....((.......)).....)))))))))..)..................((.....))..))))))................. (-13.72 = -14.12 + 0.40)

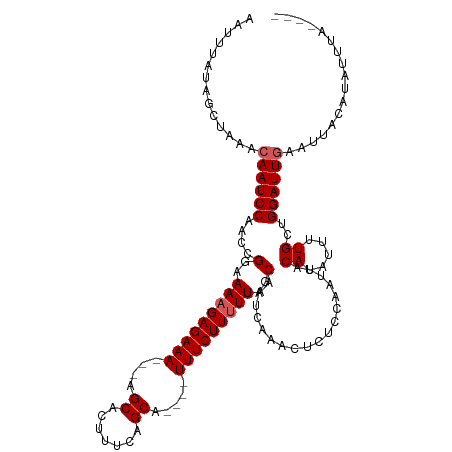
| Location | 12,563,398 – 12,563,505 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.44 |
| Shannon entropy | 0.22327 |
| G+C content | 0.32461 |
| Mean single sequence MFE | -19.84 |
| Consensus MFE | -13.08 |
| Energy contribution | -12.88 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.865060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12563398 107 - 22422827 GAAAUUCAAUUCAAACA-----AAAAAUUACUU----AGUAAUUUAUAUCUAAACAAUCCAACCGGAAAAGAGAAAAGCAGCACUUUCAGCA----UUUCUUUUUAACGAUCAAACUUUC ((...............-----..(((((((..----.)))))))..................((..(((((((((.((..........)).----)))))))))..)).))........ ( -13.20, z-score = -2.16, R) >droSim1.chrX_random 3473692 108 - 5698898 GAAAUUCAAUUCGGACA-----AAAAAUUGCUUAGUUAGUAAUUUAUAGCUAAACAAUCCAACCGGAAAAGAGAAA---AGCACUUUCAGCA----UUUCUUUUUAUCAAUCAAACUCUC ((.......(((((...-----....((((.(((((((........))))))).))))....)))))(((((((((---.((.......)).----))))))))).))............ ( -20.00, z-score = -2.99, R) >droSec1.super_21 317563 112 - 1102487 GAAAUUCAAUUCGAACA-----AAAAAUUGCUUAGUUAGUAAUUUAUAGCUAAACAAUCCAACCGGAAAAGAGAAA---AGCACUUUCAGCAUUUCUUUCUUUUUAUCAAUCAAACUCUC ............((...-----....((((.(((((((........))))))).)))).......(((((((((((---.((.......)).))))..))))))).))............ ( -15.40, z-score = -1.35, R) >droYak2.chrX 6835327 113 - 21770863 GAAAUUCGAUUCGGACAACAACAAAAAUUGCUCGGAUAGUGAUUUAUAGCCAAACAAUCCAACCGGAAAAGAGAAA---UGCACUUUCUGCA----UUUCUUUUUAACGAUCAAACUCUC .....(((.(((((....(((......)))...((((.((.............)).))))..)))))(((((((((---((((.....))))----)))))))))..))).......... ( -25.12, z-score = -2.96, R) >droEre2.scaffold_4690 4453513 108 + 18748788 GACAUUCGAUUCGGGCA-----AAAAAUUGCUCAGAUAGUGAUUUAUAGCCAAACAAUCCAACCGGAAACGAGAAA---UGCACUUUCUGCA----UUUCUUUUUAACGAUCAAACUCUC .......(((.(((((.-----..((((..((.....))..))))...)))............((....))(((((---((((.....))))----)))))......)))))........ ( -25.50, z-score = -3.06, R) >consensus GAAAUUCAAUUCGGACA_____AAAAAUUGCUUAGAUAGUAAUUUAUAGCUAAACAAUCCAACCGGAAAAGAGAAA___AGCACUUUCAGCA____UUUCUUUUUAACGAUCAAACUCUC ........................((((((((.....))))))))............(((....)))(((((((((....((.......)).....)))))))))............... (-13.08 = -12.88 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:37:56 2011