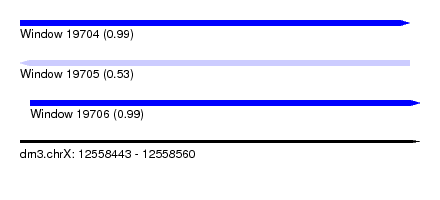
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,558,443 – 12,558,560 |
| Length | 117 |
| Max. P | 0.994539 |

| Location | 12,558,443 – 12,558,557 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 98.25 |
| Shannon entropy | 0.02417 |
| G+C content | 0.44152 |
| Mean single sequence MFE | -30.69 |
| Consensus MFE | -30.61 |
| Energy contribution | -30.39 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.78 |
| Structure conservation index | 1.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.994539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12558443 114 + 22422827 UAUUGAACCUGGCUAUAAAAAGGCUAAAGUGAGCAUAUCAACUAAACUCACUACCCAACAGGGCACUUAAACUCAUGAAAACGCACAUGAGGACAUGAGGCCGGGGCAGCAACA ..(((..(((((((.....(((.....((((((.............)))))).(((....)))..)))...((((((........)))))).......)))))))....))).. ( -30.92, z-score = -2.94, R) >droSec1.super_21 312718 114 + 1102487 UAUUGAACCUGGCUAUAAAAAGGCUAAAGUGAGCAUAUCAACUAAACUCACUAUCCAACAGGGCACUUAAACUCAUGAAAACGCACAUGAGGACAUGAGGCCGGGGCAGCAGCA .......(((((((.....(((((...((((((.............))))))..((....)))).)))...((((((........)))))).......)))))))((....)). ( -30.72, z-score = -2.43, R) >droSim1.chrX 9607201 114 + 17042790 UAUUGAACCUGGCUAUAAAAAGGCUAAAGUGAGCAUAUCAACUAAACUCACUAUCCAAAAGGGCACUUAAACUCAUGAAAACGCACAUGAGGACAUGAGGCCGGGGCAGCAACA ..(((..(((((((.....(((((...((((((.............))))))..((....)))).)))...((((((........)))))).......)))))))....))).. ( -30.42, z-score = -2.97, R) >consensus UAUUGAACCUGGCUAUAAAAAGGCUAAAGUGAGCAUAUCAACUAAACUCACUAUCCAACAGGGCACUUAAACUCAUGAAAACGCACAUGAGGACAUGAGGCCGGGGCAGCAACA ..(((..(((((((.....(((((...((((((.............))))))..((....)))).)))...((((((........)))))).......)))))))....))).. (-30.61 = -30.39 + -0.22)

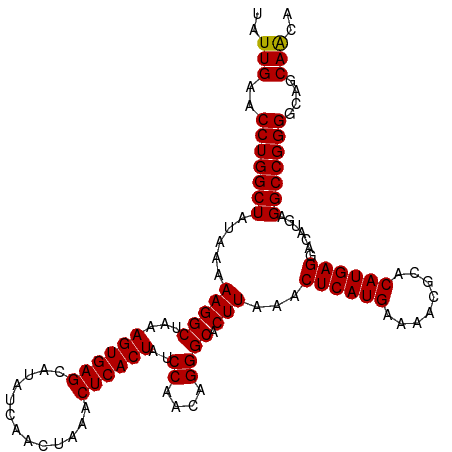
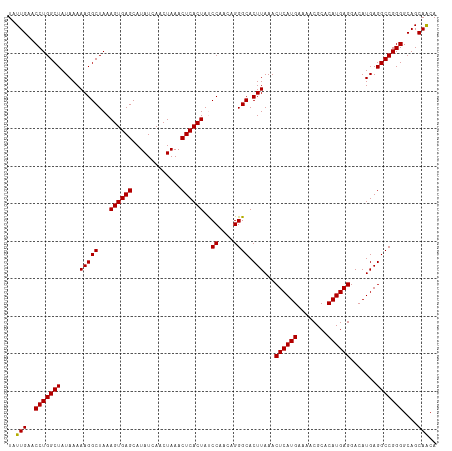
| Location | 12,558,443 – 12,558,557 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 98.25 |
| Shannon entropy | 0.02417 |
| G+C content | 0.44152 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -27.17 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.525415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12558443 114 - 22422827 UGUUGCUGCCCCGGCCUCAUGUCCUCAUGUGCGUUUUCAUGAGUUUAAGUGCCCUGUUGGGUAGUGAGUUUAGUUGAUAUGCUCACUUUAGCCUUUUUAUAGCCAGGUUCAAUA (((((..(((..(((....((..((((((........))))))..))..(((((....)))))((((((.((.....)).))))))...............))).))).))))) ( -31.90, z-score = -2.03, R) >droSec1.super_21 312718 114 - 1102487 UGCUGCUGCCCCGGCCUCAUGUCCUCAUGUGCGUUUUCAUGAGUUUAAGUGCCCUGUUGGAUAGUGAGUUUAGUUGAUAUGCUCACUUUAGCCUUUUUAUAGCCAGGUUCAAUA ..((((((....(((((..((..((((((........))))))..)))).))).....((..(((((((.((.....)).)))))))....))......))).)))........ ( -27.70, z-score = -0.62, R) >droSim1.chrX 9607201 114 - 17042790 UGUUGCUGCCCCGGCCUCAUGUCCUCAUGUGCGUUUUCAUGAGUUUAAGUGCCCUUUUGGAUAGUGAGUUUAGUUGAUAUGCUCACUUUAGCCUUUUUAUAGCCAGGUUCAAUA (((((..(((..(((....((..((((((........))))))..))...........((..(((((((.((.....)).)))))))....))........))).))).))))) ( -28.60, z-score = -1.61, R) >consensus UGUUGCUGCCCCGGCCUCAUGUCCUCAUGUGCGUUUUCAUGAGUUUAAGUGCCCUGUUGGAUAGUGAGUUUAGUUGAUAUGCUCACUUUAGCCUUUUUAUAGCCAGGUUCAAUA (((((..(((..(((....((..((((((........))))))..))((.((((....))..(((((((.((.....)).)))))))...)))).......))).))).))))) (-27.17 = -27.50 + 0.33)

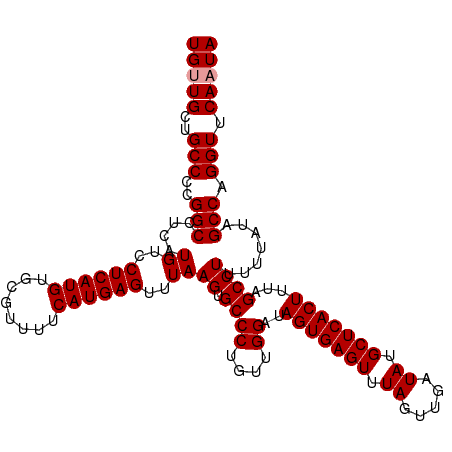
| Location | 12,558,446 – 12,558,560 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 98.25 |
| Shannon entropy | 0.02417 |
| G+C content | 0.45029 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -29.79 |
| Energy contribution | -29.79 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.993168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12558446 114 + 22422827 UGAACCUGGCUAUAAAAAGGCUAAAGUGAGCAUAUCAACUAAACUCACUACCCAACAGGGCACUUAAACUCAUGAAAACGCACAUGAGGACAUGAGGCCGGGGCAGCAACAACA ....(((((((.....(((.....((((((.............)))))).(((....)))..)))...((((((........)))))).......)))))))............ ( -30.22, z-score = -2.82, R) >droSec1.super_21 312721 114 + 1102487 UGAACCUGGCUAUAAAAAGGCUAAAGUGAGCAUAUCAACUAAACUCACUAUCCAACAGGGCACUUAAACUCAUGAAAACGCACAUGAGGACAUGAGGCCGGGGCAGCAGCAACA ....(((((((.....(((((...((((((.............))))))..((....)))).)))...((((((........)))))).......)))))))((....)).... ( -30.72, z-score = -2.53, R) >droSim1.chrX 9607204 114 + 17042790 UGAACCUGGCUAUAAAAAGGCUAAAGUGAGCAUAUCAACUAAACUCACUAUCCAAAAGGGCACUUAAACUCAUGAAAACGCACAUGAGGACAUGAGGCCGGGGCAGCAACAACA ....(((((((.....(((((...((((((.............))))))..((....)))).)))...((((((........)))))).......)))))))............ ( -29.72, z-score = -2.88, R) >consensus UGAACCUGGCUAUAAAAAGGCUAAAGUGAGCAUAUCAACUAAACUCACUAUCCAACAGGGCACUUAAACUCAUGAAAACGCACAUGAGGACAUGAGGCCGGGGCAGCAACAACA ....(((((((.....(((((...((((((.............))))))..((....)))).)))...((((((........)))))).......)))))))............ (-29.79 = -29.79 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:37:52 2011