
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,523,130 – 12,523,232 |
| Length | 102 |
| Max. P | 0.587886 |

| Location | 12,523,130 – 12,523,232 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 71.42 |
| Shannon entropy | 0.60570 |
| G+C content | 0.62481 |
| Mean single sequence MFE | -40.02 |
| Consensus MFE | -18.28 |
| Energy contribution | -18.61 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.73 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.587886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
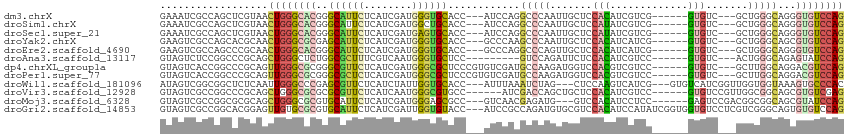
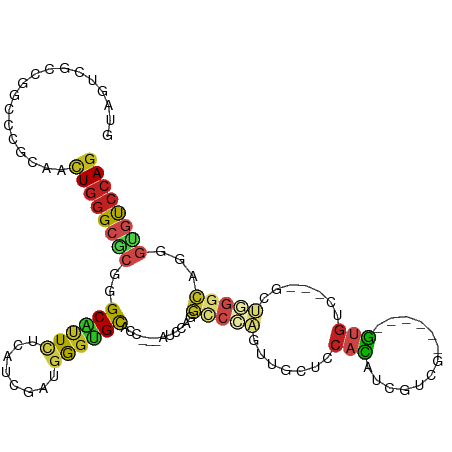
>dm3.chrX 12523130 102 + 22422827 GAAAUCGCCAGCUCGUAACUGGGCACGGGCAUUCUCAUCGAUGGGUGCACC---AUCCAGGCCCAAUUGCUCCACAUCGUCG------GUGUC---GCUGGGCAGGGUGUCCAG ..................((((((((((((((((........)))))).))---..((..(((((...((..(((.......------)))..---))))))).)))))))))) ( -38.00, z-score = -1.00, R) >droSim1.chrX 9594406 102 + 17042790 GAAAUCGCCAGCUCGUAACUGGGCACGGGCAUUCUCAUCGAUGGCUGCACC---AUCCAGGCCCAAUUGCUCCAUAUCGUCG------GUGUC---GCUGGGCAGGGUGUCCAG ..........(((((....)))))..(((((((((....(((((.....))---)))...(((((.(.((.((........)------).)).---).)))))))))))))).. ( -35.10, z-score = -0.42, R) >droSec1.super_21 277884 102 + 1102487 GAAAUCGCCAGCUCGUAACUGGGCACGGGCAUUCUCAUCGAUGAGUGCACC---AUCCAGGCCCAAUUGCUCCAUAUCGUCG------GUGUC---GCUGGGCAGGGUGUCCAG ..................((((((((((((((((........)))))).))---..((..(((((.(.((.((........)------).)).---).))))).)))))))))) ( -38.30, z-score = -1.60, R) >droYak2.chrX 6799923 102 + 21770863 GAAGUCGCCAGCACGCAACUGGGCGCGAGCAUUCUCAUCGAUGGGUGCACC---GCCCAAGCCCAAUUGCUCCACAUCAUCG------GUGUC---GCUGGGCAGCGUGUCCAG ..........((((((...((((((.(.((((((........)))))).))---))))).(((((...((..(((.......------)))..---))))))).)))))).... ( -42.40, z-score = -2.06, R) >droEre2.scaffold_4690 4412490 102 - 18748788 GAAGUCGCCAGCCCGCAACUGGGCACGGGCAUUCUCAUCGAUGGGUGCACC---GCCCAGGCCCAGUUGCUCCACAUCAUCG------GUGUC---GCUGGGCAGGGUGUCCAG ...(.((((.((((((((((((((.(((((((((........)))))).))---).....))))))))))...((((.....------)))).---...))))..)))).)... ( -48.00, z-score = -2.36, R) >droAna3.scaffold_13117 3430443 96 + 5790199 GUAGUCUCCGGCCCGCAGCUGGGCUCUGGCGCUUUCGUCAAUGGGUGCUCC---------GUCCAGAUUCUCCACAUCGUCC------GUGUC---ACUGGGCAGAGUAUCCAG ..((..((((((.....))))))..))((((....))))..(((((((((.---------((((((......(((.......------)))..---.)))))).))))))))). ( -35.80, z-score = -1.85, R) >dp4.chrXL_group1a 3308203 105 + 9151740 GUAGUCACCGGCCCGCAGUUGGGCGCGGGCGUUCUCAUCGAUGGGCGCUCCCGUGUCGAUGCCAAGAUGGUCCACGUCGUCC------GUGUC---GCUUGGCAGGACGUCCAG ..........(((((....)))))..(((((((((.(((((((((....)))..))))))((((((.(((..((((.....)------)))))---)))))))))))))))).. ( -44.10, z-score = -0.62, R) >droPer1.super_77 55845 105 - 256778 GUAGUCACCGGCCCGCAGUUGGGCGCGGGCGCUCUCAUCGAUGGGCGCUCCCGUGUCGAUGCCAAGAUGGUCCACGUCGUCC------GUGUC---GCUUGGCAGGACGUCCAG ...(((..(((((((....)))))(((((.((.((((....)))).)).)))))..)).(((((((.(((..((((.....)------)))))---)))))))).)))...... ( -42.60, z-score = 0.16, R) >droWil1.scaffold_181096 1883269 105 + 12416693 AUAGUCGGCGGCUCUCAAUUGGGCCCGAGCGUUCUCAUCUAUUGGUGCACC---AUUUAAAUCUAG---CUCCAAGUCAUCG---GUGUCAUCGGUUGGUGGUAAAGUGCCCAC ...((.((((((((......)))))...((.((..((((.(((((((((((---............---............)---))).))))))).))))..)).))))).)) ( -29.26, z-score = -0.71, R) >droVir3.scaffold_12928 1106510 102 + 7717345 GUAGUCGCCGGCCCGCAGCUGGGCGCGCGCGUUCUCAUCAAUGGGCGUGCC------AUCGACCAGCUGCUCCACAUCGUCC------GUGUCCGUUGGCGGCAGCGUGUCGAG ((.(((((((((..((((((((.((.((((((((........)))))))).------..)).))))))))..(((.......------)))...))))))))).))........ ( -47.80, z-score = -1.70, R) >droMoj3.scaffold_6328 2086080 102 + 4453435 GUAGUCGCCGGCGCGCAGCUGGGCGCGUGCAUUCUCAUCGAUGGGAGCGCC---GUCAACGAGAUG---GUCCACAUCCUCC------GAGUCCGACGGCGGCAGCGUAUCCAG ((.(((((((((((((.((...))))))))...(((...((((((...(((---(((.....))))---)))).))))....------))).....))))))).))........ ( -41.00, z-score = -0.14, R) >droGri2.scaffold_14853 2701345 111 - 10151454 GUAGUCGCCGGCACGGAGUUGUGCGCGUGCAUUCUCAUCGAUUGGUGUACC---AUCCGCCAGAUGUGCGUCCACAUCCAUAUCGGUGGUGUCCUCGUCGGGCAGUGUGUCCAG ....((((((..(.(((..((((.(((..(((.((...((..(((....))---)..))..)))))..))).))))))).)..))))))(((((.....))))).......... ( -37.90, z-score = -0.04, R) >consensus GUAGUCGCCGGCCCGCAACUGGGCGCGGGCAUUCUCAUCGAUGGGUGCACC___AUCCAGGCCCAGUUGCUCCACAUCGUCG______GUGUC___GCUGGGCAGGGUGUCCAG ..................((((((((..((((((........))))))............(((((.......(((.............))).......)))))...)))))))) (-18.28 = -18.61 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:37:47 2011