| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,475,774 – 12,475,865 |
| Length | 91 |
| Max. P | 0.778791 |

| Location | 12,475,774 – 12,475,865 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 58.13 |
| Shannon entropy | 0.84300 |
| G+C content | 0.69138 |
| Mean single sequence MFE | -42.16 |
| Consensus MFE | -17.20 |
| Energy contribution | -16.66 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 2.09 |
| Mean z-score | -0.70 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.778791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 12475774 91 - 22422827 -AGUCGGGACACGGCCUGGCGCCA--GAUGGCAGUGCUGGCAACUCC---CUAUCCGCCGGCGGAGGCGUAUCCGGCGGUUGUCCGCCCGGCAUCAA -.(((((((.(((.(((...(((.--...)))..(((((((......---......))))))).)))))).)))(((((....)))))))))..... ( -41.20, z-score = -0.13, R) >droSim1.chr3R 13581219 83 + 27517382 ------------CGCGUGAGGACGGAUUGUUCCUGGUUCGCGAGUCCACAAACUUCCCUGGCGACUACACGCUCUGCGUUUGUU--UCCAGUCCAAG ------------.......(((((((..(((..(((..(....).)))..))).))).(((.(((...((((...))))..)))--.)))))))... ( -24.50, z-score = -0.50, R) >droSec1.super_21 230709 91 - 1102487 -AGUCGGGGCACGGCCUGCCGCCA--GAUGGCGGCUCUGGCAACUCC---CUAUCCGCCGGCGGAGGCGUGUCCGGAGGCGGUCCGCCCGGCAUCAA -.(((((((.((.(((.((((((.--...))))))(((((..((.((---...((((....)))))).))..)))))))).)).).))))))..... ( -48.00, z-score = -0.89, R) >droYak2.chrX 6751536 91 - 21770863 -AAUCGGGACACGGCCUGCCGCCA--GAUGGCGGGGCUGGCAACGCC---CUAUCCGCCGGCGGAGGCGUGGCCGGUGGCGGUCCGCCCGGCAUCAA -..(((((..((.(((..(((((.--...))))).((((((.(((((---...((((....))))))))).))))))))).))...)))))...... ( -50.60, z-score = -0.93, R) >droEre2.scaffold_4690 4364934 91 + 18748788 -AAUCGGGACACGGCCUGCCGCCA--GAUGGCGGUCCUGGCAACGCC---CUAUCCGCCGGCGGAGGCGUGGCCGGUGGCGGUCCGCCCGGCAUCAA -..(((((..((.(((.((((((.--...)))))).(((((.(((((---...((((....))))))))).))))).))).))...)))))...... ( -49.20, z-score = -1.05, R) >droAna3.scaffold_13117 3366349 84 - 5790199 -----UGCACAUGGCAUACCGCCG--GAUAAUGCGGC--GCCACUGU---C-GGCUGGAGUUGGAGGAGUUGGCGGCAAUGGUCCGUCCGGCGUUAA -----(((.....)))...(((((--(.....((((.--((((.(((---(-(.(..(..(.....)..)..)))))).)))))))))))))).... ( -31.00, z-score = 0.37, R) >droMoj3.scaffold_6328 2022924 97 - 4453435 CGCACGGGCCACCGCACGUUGGCGUCGGCGUCGGCGUCAGCGUUGCCGAUCUCGCUGCCGGCAGCAGCAGCGUCGGCGUCGGCCCCUCCAGCGUCAA (((..(((((.....((((((((((((....)))))))))))).((((((((.(((((.....))))))).))))))...))))).....))).... ( -50.60, z-score = -1.75, R) >consensus _A_UCGGGACACGGCCUGCCGCCA__GAUGGCGGGGCUGGCAACUCC___CUAUCCGCCGGCGGAGGCGUGGCCGGCGGCGGUCCGCCCGGCAUCAA ......((.....(((..((((((....))))))....)))............((((....)))).......))((((.(((.....))).)))).. (-17.20 = -16.66 + -0.54)

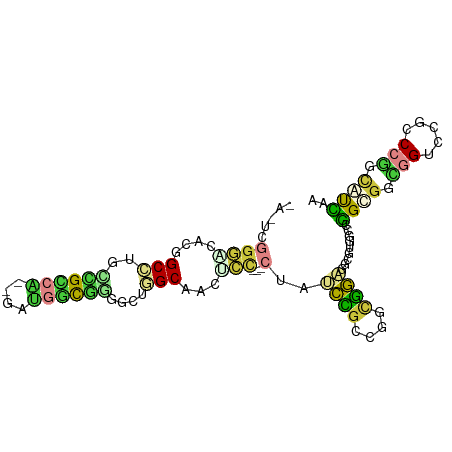

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:37:44 2011