
| Sequence ID | dm3.chrX |
|---|---|
| Location | 12,455,661 – 12,455,734 |
| Length | 73 |
| Max. P | 0.872097 |

| Location | 12,455,661 – 12,455,734 |
|---|---|
| Length | 73 |
| Sequences | 9 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 70.19 |
| Shannon entropy | 0.62516 |
| G+C content | 0.51403 |
| Mean single sequence MFE | -16.38 |
| Consensus MFE | -8.84 |
| Energy contribution | -9.29 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.872097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
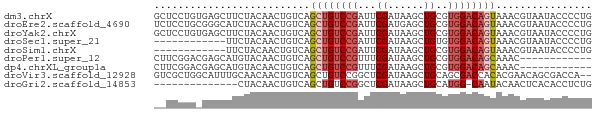
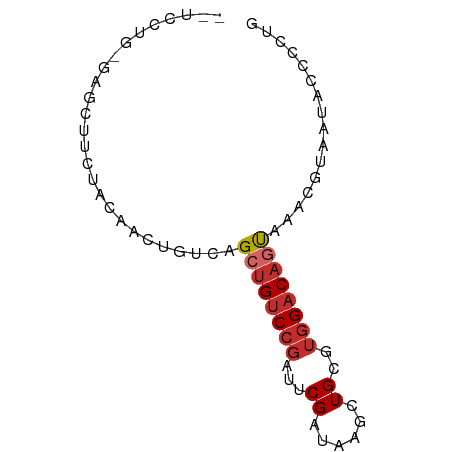
>dm3.chrX 12455661 73 - 22422827 GCUCCUGUGAGCUUCUACAACUGUCAGCUGUCCGAUUCGAUAAGCUGCGUGGACAGUAAACGUAAUACCCCUG ((((....))))..........((..((((((((...((......))..))))))))..))............ ( -16.50, z-score = -0.92, R) >droEre2.scaffold_4690 4344513 73 + 18748788 UCUCCUGCGGGCAUCUACAACUGUCAGCUGUCCGAUUCGAUGAGCUGCGUGGACAGUAAACGUAAUACCCCUG .....((((((((........)))).((((((((...((......))..))))))))...))))......... ( -17.10, z-score = -0.14, R) >droYak2.chrX 6731724 73 - 21770863 GCUCCUGUGAGCUUCUACAACUGUCAGCUGUCCGAUUCGAUAAGCUGCGUGGACAGUAAACGUAAUACCCCUG ((((....))))..........((..((((((((...((......))..))))))))..))............ ( -16.50, z-score = -0.92, R) >droSec1.super_21 210977 61 - 1102487 ------------UUCUACAACUGUCAGCUGUCCGAUUCGAUAAGCUGUGUGGACAGUAAACGUAAUACCCCUG ------------..........((..((((((((...((......))..))))))))..))............ ( -12.90, z-score = -1.29, R) >droSim1.chrX 9574416 61 - 17042790 ------------UUCUACAACUGUCAGCUGUCCGAUUCGAUAAGCUGCGUGGACAGUAAACGUAAUACCCCUG ------------..........((..((((((((...((......))..))))))))..))............ ( -12.20, z-score = -0.97, R) >droPer1.super_12 2358586 61 + 2414086 CUUCGGACGAGCAUGUACAACUGUCAGCUGUCCGUUUCGAUAAGCUGCGUGGACAGCAAAC------------ ...(((((.(((.((.((....)))))))))))).........((((......))))....------------ ( -17.40, z-score = -0.83, R) >dp4.chrXL_group1a 3246983 61 - 9151740 CUUCGGACGAGCAUGUACAACUGUCAGCUGUCCGUUUCGAUAAGCUGCGUGGACAGCAAAC------------ ...(((((.(((.((.((....)))))))))))).........((((......))))....------------ ( -17.40, z-score = -0.83, R) >droVir3.scaffold_12928 1020045 71 - 7717345 GUCGCUGGCAUUUGCAACAACUGUCAGCUGUCCGGCUCGAUAAGCUGCAGCGACCACACGAACAGCGACCA-- (((((((((....)).......(((.(((((..((((.....))))))))))))........)))))))..-- ( -26.40, z-score = -2.28, R) >droGri2.scaffold_14853 2620340 58 + 10151454 --------------CUACAACUGUCAGCUGUCCGGCUCGAUAAGCUGCAUGG-CAAUACAACUCACACCUCUG --------------............(((((.(((((.....))))).))))-)................... ( -11.00, z-score = -0.69, R) >consensus __UCCUG_GAGCUUCUACAACUGUCAGCUGUCCGAUUCGAUAAGCUGCGUGGACAGUAAACGUAAUACCCCUG ..........................((((((((...((......))..))))))))................ ( -8.84 = -9.29 + 0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:37:42 2011